RESEARCH ARTICLE Open Access Convergence Of Logic Of .
Kravchenko-Balasha et al. BMC Systems Biology 2011, EARCH ARTICLEOpen AccessConvergence of Logic of Cellular Regulation inDifferent Premalignant Cells by an InformationTheoretic ApproachNataly Kravchenko-Balasha1†, F Remacle2,3†, Ayelet Gross3, Varda Rotter4*, Alexander Levitzki1* and RD Levine3,5*AbstractBackground: Surprisal analysis is a thermodynamic-like molecular level approach that identifies biologicalconstraints that prevents the entropy from reaching its maximum. To examine the significance of altered geneexpression levels in tumorigenesis we apply surprisal analysis to the WI-38 model through its precancerous states.The constraints identified by the analysis are transcription patterns underlying the process of transformation. Eachpattern highlights the role of a group of genes that act coherently to define a transformed phenotype.Results: We identify a major transcription pattern that represents a contraction of signaling networks accompaniedby induction of cellular proliferation and protein metabolism, which is essential for full transformation. In addition,a more minor, “tumor signature” transcription pattern completes the transformation process. The variation withtime of the importance of each transcription pattern is determined. Midway through the transformation, at thestage when cells switch from slow to fast growth rate, the major transcription pattern undergoes a total inversionof its weight while the more minor pattern does not contribute before that stage.Conclusions: A similar network reorganization occurs in two very different cellular transformation models: WI-38and the cervical cancer HF1 models. Our results suggest that despite differences in a list of transcripts expressed indifferent cancer models the rationale of the network reorganization remains essentially the same.BackgroundDeciphering regulatory events that drive malignanttransformation represents a major challenge for systemsbiology. Here, we analyzed the genome-wide transcription profiling of an in vitro cellular system, in whichcells were induced to transform to a cancerous phenotype, through intermediate states. Cells evolving towardsa malignant state exhibit changes in gene expressionthat do away with pathways that interfere with proliferation [1]. Cancer cells also appear to be less subject tosome of the restrictions and controls characteristic ofmulticellular organisms [1]. For different cancers many* Correspondence: varda.rotter@weizmann.ac.il; � Contributed equally1Unit of Cellular Signaling, Department of Biological Chemistry, TheAlexander Silberman Institute of Life Sciences, The Hebrew University ofJerusalem, Jerusalem 91904, Israel3The Fritz Haber Research Center for Molecular Dynamics, The Institute ofChemistry, The Hebrew University of Jerusalem, Jerusalem 91904, IsraelFull list of author information is available at the end of the articleoncogenes and tumor suppressors have been identified[2], but determining a list of genes that characterizecancers has not been fully successful [3].In this study we are using a physically motivatedmethod of gene expression analysis based on the proposition that the process of gene expression is subject tothe same quantitative laws as inanimate nonequilibriumsystems in physics and chemistry. This allows us toapply a thermodynamic-like approach where entropy isa physical quantity and not only a statistical measure ofdispersion. Our purpose is similar to earlier studies ofgroupings of genes [4,5] including the validation [6] ofthe predicted [4,5] mechanism of regulation. The papersof Janes et al [7-9] are close to our aims as the implementation of co-clustering methods to detect similarexpression patterns, e.g., [10]. The maximal entropymethod has been used to identify association of genes[11,12]. We too assume that the entropy depends on thedistribution of species. The essential difference is that inour case entropy is not just the mixing entropy. This is 2011 Kravchenko-Balasha et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of theCreative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, andreproduction in any medium, provided the original work is properly cited.
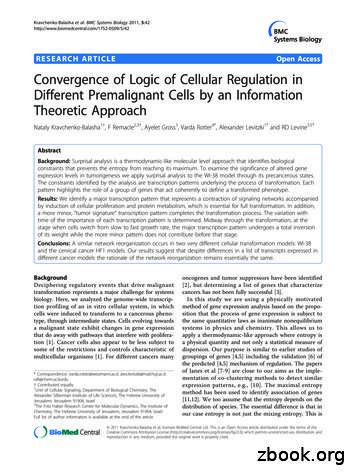
Kravchenko-Balasha et al. BMC Systems Biology 2011, ause the value of the thermodynamic entropydepends on the distribution of species and on the internal structure of each. The result is that at the maximumof the entropy our distribution is not uniform. Ourwork differs from Boolean based approaches [13] wherea gene is either expressed or not. Probabilistic networks[14-18] are closer in that they determine a kinetic orderin time. Time series data is often analyzed using principal component analysis or partial least squares regression, e.g., Janes et al [7-9]. Implementing surprisalanalysis of a high throughput data set is convenientlycarried out by diagonalizing a covariance matrix. But itis the covariance matrix of the logarithm of the expression levels and this means that the levels need not bemean centered prior to the diagonalization.The information-theoretic analysis that we use iscalled surprisal analysis [19] to emphasize that at maximal entropy genes are not necessarily equally expressed.In each stage of development, the transient gene expression patterns and their associated biological phenotypesare identified as constraints that prevent the entropyfrom achieving the maximal possible value. The theoryis thermodynamics-like because it also invokes the timeinvariant distribution of expression levels. We show howto determine this distribution from the data and findthat it is not necessarily uniform. This is to be expectedbecause this steady distribution reflects the free energyof the mRNA molecules.The biggest extent of deviation from the maximalentropy defines the major transcription pattern thatoccurs during the process of transformation. We alsodefine minor transcription patterns that participate inthe establishment of cancer. The major pattern isimportant throughout while more minor patterns typically contribute significantly only early or only later on.We will specifically emphasize the stages during the cellular transformation when the role of an expression pattern undergoes an ‘inversion’ in its significance. By‘inversion’ we refer to a time course where genes thatwere highly expressed at the stage before, undergo achange to being under expressed in the stage after andvice versa. A model [20,21] where different processesare initiated, some that eventually lead to malignancyand some that do not, is analyzed in detail to illustratethese ideas. Several of the processes initiated in themodel system share a common earlier stage. At laterstages the formalism is able to point out the differencesthat evolve from those initially common patterns.The technical mathematical details are spelled out inthe Additional file 1 online. In practical terms theresults of the analysis is a ranking of the gene expression patterns according to the importance of their contribution at each stage. In the Additional file 1 thenotion of the ‘importance’ of a pattern is defined andPage 2 of 14quantified. Using the ‘importance’ we show below that arather small number of expression patterns suffice toquantitatively reproduce the expression levels of all individual genes. One or two of the most important patternsalready provide a close characterization of the expression levels.We analyze the changes in the gene expression levelsduring the process of carcinogenesis in the thoroughlystudied cellular model WI-38, developed by one of us[10,20,21]. The cancer model system follows the progression from the normal phenotype all the way to theonset of cancer [20,21]. The WI-38 cellular systemincludes parental WI-38 fibroblasts in the young, senescent stages as well as the hTERT immortalized cells atthe different stages [20,21]. At a certain stage (Figure 1),p53 was inactivated by the expression of a dominantnegative peptide GSE56 [20,21], and the expression ofoncogenic H-Ras was induced by infection at the indicated time points as shown in Figure 1[20,21]. Thegenetic alterations were applied at different points asshown in Figure 1 where the time points are labeledT 1,2,.,12. It is important to note that different trajectories of the transformation process go through differenttime points. For example, we will compare the three trajectories 1-5-7-8-9, 1-5-7-8-11 and 1-5-7-8-10-12, whichshare a common process up to and including point 8.These are all continuous processes where each timepoint follows the preceding one and we will refer tosuch a sequence of stages as a trajectory. An oppositeexample is the trajectory 1-5-6-7 that cannot be considered as continuous since time point 7 does not experimentally follow point 6.The novelty and a power of our approach lies in ourability to identify the major and minor gene expressionpatterns in each stage ( time point) during the transformation. Moreover this analysis identifies the necessary and sufficient transcription patterns that define thecellular transformation. Additionally our analysis identifies new networks that participate and contribute significantly to the establishment of the different phenotypesduring the transformation. The patterns identified bythe present study are further examined by comparisonto the results of the original analysis of the WI-38 system [10,20,21], see also Additional file 1. Furthermore,our analysis considers different trajectories that have different outcomes, depending on which perturbationswere applied. For example, trajectory 1-5-7-8-11 has adifferent outcome from trajectory 1-5-7-8-9 as can beseen in Figure 1.The model developed in the Rotter lab uses fibroblastswhile in an earlier recent study [22,23] we used HPV-16immortalized keratinocytes. Moreover, the Rotter model(Figure 1) differs markedly in the route of transformation. Even so, we find a convergence of the dominant
Kravchenko-Balasha et al. BMC Systems Biology 2011, e 3 of 14H-Rasp53610p53p53875p53hTERTCancer in12 nude sp53404140215355490Figure 1 Schematic representation of the WI-38 cell model (adapted from [21]). Schematic representation of the physiological state (young,senescent, immortal, tumorigenic) and introduced modifications (hTERT, H-RAS, p53 inactivation) of the WI-38 cells along the process ofmalignant transformation. The stages chosen for the theoretical analyses can be arranged as several continuous trajectories where each samplefollows the preceding one. A common route for many trajectories, the ones we highlight in the text, is represented by the blue color. The firstbranching occurs at the point 6 (pale blue) and generates the trajectory 156. The second branching occurs at the point 8 and generates 3trajectories: 1-5-7-8-10-12 (red), 1-5-7-8-11 (yellow) and 1-5-7-8-9 (purple). There is an additional independent trajectory 1-3-4 (black). PDLs are thenumber of doublings since the cells primary isolation in vitro.expression patterns and we identify a similar rationalebehind the process of carcinogenesis. This recognitionof a common rationale is a key result of our work. Wesuggest that the underlying principle of transcriptionnetwork reorganization is common to the different cancer cell models.The presentation of the results follows two lines. Oneis the discussion of the information theoretic based tool,which utilizes gene expression levels to identify transcription patterns and to determine their contributionto the cancer transformation process at each stage. Thecomplementary development is the biological interpretation of the calculated patterns and their weights.ResultsTheoretical Section: The information theoretic analysisThis section summarizes the essence of the informationtheory based method used for the analysis of mRNAarray as described in detail in the Additional file 1 section “Surprisal analysis”. For additional discussion of themotivation, see [23]. References 24-28 provide morebackground. Here we just emphasize that in general ourapproach, known as surprisal analysis, [19] is a methodof analysis of systems in both equilibrium and not equilibrium that are subject to constraints. Surprisal analysisis an analysis of the logarithm of the expression level ofeach gene as in equation (1) below. This analysis determines the transcription patterns of the transformationprocess and the weights of these independent patternsat each stage ( time point) of the transformation.A transcription pattern is a set of transcripts that actcoherently. We index the patterns by the Greek letter a,a 1,2,. label the different independent patterns. Foreach pattern we determine its importance. la(tT) is thevalue ( the importance) of the contribution of the pattern a at time point T. We shall show that at any stagethere are very few important patterns. The validation ofthis statement as well as the determination of the transcription patterns is quantitative. The information theoretic thermodynamic-like approach derives the
Kravchenko-Balasha et al. BMC Systems Biology 2011, e 4 of 14logarithm of the expression level of each gene. For genei at the time point T we obtain equation (1) for theexpression level of gene i at the time point T where Giais the weight of that gene in the pattern a ln Xi (tT )Giα λα (tT ) ln Xo i α 1 measuredexpression level of gene ideviation due to phenotypesexpression level of gene iat a steady stateof the transformation process(1)The first term on the right side of the equation is thetime-invariant part of the gene expression level for theparticular transformation process. Typically this termmakes a non-negligible contribution. According to thetheory, this term is the gene expression level at maximalentropy. The time varying transcription patterns arerepresented by the terms in the sum. It is these termsthat reduce the magnitude of the entropy. In the information theory approach la(tT) is the extent of reductionof the entropy due to the particular gene transcriptionpattern a. Due to the presence of the constraints, represented in equation (1) by the action of expressed genes,the system does not reach a steady state.We have an accurate representation of the transformation process when the measured left hand side in equation (1) is close in value to the theoretical representationon the right hand side of equation (1). By making a leastsquares match between the two sides of equation (1) weobtain the numbers G ia and l a (t) with the necessarymathematical details provided in the Additional file 1with background material provided in [24-28]. As alreadymentioned, only very few terms in the sum in equation(1) are required to achieve this. The mathematical technique we use insures that the patterns are orthogonal andindependent. We do not seek a perfect fit because experimental data is invariably accompanied by some noise.Theoretical Section: Implementation of Surprisal analysisBy the end of the thermodynamic-like analysis weassociate the deviations from steady state with a set oftranscription patterns. Note that in our approach, eachpattern is permanent and not varying in time. The listof coefficients Gia is determined by our analysis for eachvalue of a, see Additional file 1 and [23] for details. Theweights Gia are not a function of time. Only the weightla(tT) of the transcription pattern varies with time. Thisis the background necessary for the analysis to be implemented below. We next proceed to make some additional points about the theory.A technical point is that the theory expresses theweight of a pattern, that is its importance, as a productof two factors, la(tT) ωaPaT, see the Additional file 1of this paper as well as [23]. Here ωa is the time independent weight of transcription pattern a while PaT isthe fractional weight of the contribution of pattern a attime T. (The fractional weights sums up to unity as 2 1PαT). We are interested both in those transcription patterns with a large value of the absolute weightω a and in those patterns whose fractional weightchanges significantly in the course of time. The factorization of la(tT) is not just a technical matter because itshows that a transcription pattern can have a lowerabsolute weight ωa yet its time-dependent weight canchange significantly at some stage of the transformation.The time invariant part is computed as that part of Xi(tT) that is not dependent on time. For notational reasons it is convenient to introduce a ‘zeroth’ pattern bywriting the steady state term as ln Xio Gi0 λ0 . Unlikethe other l’s, from its definition l0 does not depend ontime. In our computation we allow l0 to vary and useits theoretical constancy as a check and a numericalvalidation of the results. In the experiments of Rotteret al, [20,21] there are several distinct trajectories thatdiffer by which mutation was induced in the system atthe last point in time. Because different trajectories canterminate at distinct biological fates, each such trajectory can possess its own time-invariant pattern.In the Additional file 1 we provide full details on howthe numerical values of the weights la (t T) and of thetranscription patters Gia are determined from the measured values of the expression levels Xi(tT) of differentgenes, where i is an index of a gene, at different timestT. It follows from that technical discussion that there isan upper value for the number of different transcriptionpatterns that can be identified from the data.The result (1) was first derived in the context of selectivity of energy requirements and specificity of energydisposal of chemical reactions [25,28]. Using this expression to fit the observed data is often known as surprisalanalysis. The term surprisal refers to the informationprovided by the deviation of the expression level fromthe time independent part.The transcription patterns and constraints are identified by fitting equation (1) to the observed expressionlevels at different times along a particular trajectory. Saythat there are A time points in that trajectory. When weuse all A transcription patterns then the informationtheoretic expression (1) with a 1,2,., A-1 is an exactrepresentation of the data, so at any time point the A,l’s, l0,l1,.,lA-1 fully suffice to recover the data, noiseand all. The surprising result is that, as we shall see, inpractice one major transcription pattern often accountsfor the measured expression levels, (see Figure 2). Whatit means is that transcription patterns can be ranked interms of their importance. The details of the fitting procedure are described in the Additional file 1.The functional form (1) is derived by imposing constraints that prevent the entropy of the distributions ofgene expressions from being fully maximal. The detailsT
Kravchenko-Balasha et al. BMC Systems Biology 2011, e 5 of 14Figure 2 A scatter plot of the computed gene expressionlevels vs. the measured values. A scatter plot of the computedgene expression levels, lnXi(tT), equation (1), for 5582 transcripts attime 7 of the trajectory 15781012 vs. the measured values. Dots (redonline) computed by equation (1), using only the most dominanttranscription pattern, a 1. Squares (blue online) computed byequation (1), using the two leading transcription patterns, a 1,2.Straight line: a perfect correlation using all the five transcriptionpatterns. For this trajectory and a few others the expression levels atlate times are best accounted for using patterns 1 and 3 becausethe second pattern is far less important, see Figure 3. For certaintrajectories at early times, pattern 4 is important.are provided in [23] and in reviews of the formalism[24-27]. Technically the constraints are imposed usingthe method of Lagrange’s undetermined multipliers [29].A multiplier la(tT) is associated with each constraint aat each time point T. The value of the multiplier isdetermined by the value of the constraint at that timeT. We here determine the value by fitting equation (1)to the data and we interpret l a (t T ), the value of themultiplier a at time T, as the contribution of transcription pattern a at that time. We make the least square fitof the experimentally measured right hand side to thetheory derived left hand side of equation (1) using thematrix technique of SVD. This provides a set conjugateeigenvectors that define both the weights Gia of gene iin pattern a and the weight la (T) of the pattern a atthe time T. The distinct eigenvectors are orthogonal andthis insures the independence of the patterns.This interpretation l a (T) is directly seen when wecompute the change in gene expression between twotime points T and T’ ln Xi (tT ) ln Xi (tT ) Giα λα (tT ) λα (tT )α 1 Giα ωα (PαT PαT ) α 1(2)Equation (2) plays a key role in the quantitative evaluation of the biological implications of the results ofsurprisal analysis as reported below. Specifically, equation (2) highlights the quantitative aspects of changes inthe levels of gene expressions. Changes in expressionpatterns primarily require that the fractional weight PaTchanges significantly but it helps that the absoluteweight ωa is large. Also worth noting is that the changesin the fractional weights and in the absolute weightappear in the exponents of the levels of gene expressions. Particularly when the fractional weights changesign, see Figure 3 below, the levels of gene expressionscan change by orders of magnitude. This is part of whatwe mean by an ‘inversion’ of the level of gene expressions. An example of an inversion is shown in Figure 4below.The Additional file 1 shows how to use equation (2)to compute the entropy of the gene transcription systemat different points in time. Entropy is a state functionmeaning that it depends only on the current geneexpression levels and not on how we arrived at thesevalues. The equations given in the Additional file 1 provide an explicit illustration of this important property.Figure 3 The temporal changes in the three most importanttranscription patterns. The weights of the three most importanttranscription patterns for the trajectory that goes through the timepoints 1,5,7,8,10 and 12 see Figure 1. Digital values for alltranscription patterns and all trajectories are given in the Additionalfile 1 Table S1. The representation la(tT) ωaPaT and the plots ofthe PaT’s vs. T, Additional file 1 Figure S3 and S4, show that, forexample, pattern 3 does not contribute meaningfully at the earliertimes. The small value at early times is because, as the numericalvalue of the label aμμμincreases, the eigenvectors, see Additionalfile 1PaT are more sensitive to noise.
Kravchenko-Balasha et al. BMC Systems Biology 2011, ure 4 An inversion in the expression level genes at twoconsecutive time points. An inversion in the expression level ofone hundred genes at two consecutive points in time along thetrajectory 1-5-7-8-10-12. Only a hundred values are shown for claritybut the same pattern recurs for all values of the gene index i. Thechange in the expression level, equation (2), is a sum over allpatterns. The plot shows only the dominant, see Figure 3, term a 1, between points 7, red online, and 8, blue online. Note the ln ofthe gene level changes by a factor of more than 5, exp(5) 150. Sothe inversion is fairly extreme being by more than two orders ofmagnitude. As seen in Figure 3 there is a big change in the sign ofla 1(tT) between T 7 and 8.Theoretical Section: Application of Surprisal analysisIn this study we examined the WI-38/hTERT cell system, which was guided to develop into a fully transformed cell, beginning with the normal WI-38immortalized non-transformed fibroblasts. Cells underwent molecular manipulation such as hTERT insertion,many doublings, depression of p53 function and theinsertion of oncogenic H-ras as depicted in Figure 1.The model system was studied using the Human Genome Focus Array (Affymetrix, Santa Clara, CA) with8500 verified human genes [10,21]. The data is the geneexpression level for each transcript, Xi(tT), at time t Tfor a series of 12 time points as shown in Figure 1. Theprevious analysis [10,21] of the data identified manytransformation hallmarks. In particular down-regulationof the transcripts involved in cell development and differentiation at the early stages of transformation, induction of protein biosynthetic pathways, alteration inembryonic antigen expression and in apoptotic transcripts at the latter stages of transformation. In addition,a “tumor-forming” genetic signature reflected in highgene expression levels for cytokines and chemokineswas identified. The major findings of this previous studyPage 6 of 14were used to validate the information- theoreticalapproach used in the current analysis as discussed indetail in the Additional file 1. In order to examinewhich biological processes were most affected by thetransformation, we used the EASE software [30] togroup those transcripts that passed the t-test analysisand that changed by at least exp( 0.5) for each transcription pattern a between two time points. Biologicalcategories that were significantly over-represented, asdefined by EASE score 0.05 are shown in the Additional file 1, Tables S2 to S19.Since this system did not develop continuously fromone point to the next we divided it into several trajectories representing the various possible processes. Theexpression levels were measured in duplicates for eachtime point in the trajectory (Figure 1). The data that weanalyzed was the mean of the duplicates and included5582 genes that had a ‘present call’ (according to Affymetrix calling procedure) in the two duplicates of atleast one sample [10,21]. We also performed the analysisusing only those genes that were further filtered by therequirement that the variation between the duplicates isquite small (below 0.05 as judged by a paired t-test).Information-theoretic results of Surprisal Analysis of geneexpressionUsing the data reported by Milyavsky et al. [21], weimplemented surprisal analysis and present some resultsof the analysis in Figure 3. la(tT) represents the importance of the contribution, of gene expression pattern aat the time T. The trajectory 1-5-7-8-10-12 (Figure 1),includes 6 time points and therefore a maximum sixvalues of l a (t T ) can be calculated where a 0,1,.,5.The a 0 term is the time invariant gene expressionpattern term and the five other are the varying patternsand we rank them in order of decreasing weight.Thereby, consecutive terms in the sum of terms inequation 1 make decreasing contribution. Figure 3shows three curves that are the values of la(tT) for the3 constraints (or gene expression patterns) contributingmost to the process of transformation, as a function oftime.The dominant transcription pattern a 1 shown inFigure 3 undergoes a large change in value, accompanied by a change of the sign of its weight, between timepoints 7 and 8. The second pattern increases significantly between time points 5 and 7 and then drops tozero at point 8 and stays zero thereafter. The third pattern contributes only at the last three time points andthe sign of its value changes between points 8 and 12.As seen in Figure 3 a weight of ‘zero’ is not exactlyzero. This point is best discussed using the representation l a (t T ) ω a P aT . A weight of near zero at somevalues of time means that while the absolute weight ωa
Kravchenko-Balasha et al. BMC Systems Biology 2011, 5:42http://www.biomedcentral.com/1752-0509/5/42is not necessarily small, the fractional weight P aT issmall at those time points. (For each patternfrac a the2tional weights are normalized to one asT PαT 1 ).The weights for patterns a 1 and 2 corresponding toFigure 3 are shown in the Additional file 1 Figures S3,S4 and S5. Some patterns do not have a large weight.The theory states that a gene expression pattern doesnot contribute, meaning la(tT) 0, at such time pointsthat its presence does not lower the entropy. From thepoint of view of the expression levels of genes, a patternwith a very low weight does not contribute to the geneexpression levels at that time, see equation (1) and Figure 5 below. In this case patterns with higher weightwill contribute to the measured expression network.As shown in the Additional file 1 Table S1 the ‘zeroth’multiplier l 0 (t T ) does not depend on the time t T ofmeasurement. This value should be constant, since l0(tT) is the measure of the contribution of the time invariant gene expression pattern of the steady state, a 0.The nearly exact constancy of the fitted value of l0(tT),at different times, is a validation of the concept of atime-invariant contribution, see equation (1). a 0 isthe pattern at the maximum of the entropy withouttime dependent constraints. It is the expression patternat the limit of steady state. As expected from basic considerations, not all trajectories lead to the same cell fate.Therefore, different trajectories have different secularfates and can therefore differ in their l0 values.We rank the varying transcription patterns by theirimportance with a 1 being the dominant one whereimportance is by the size of the absolute weight ωa. Thesmaller the value of ωa, the more likely it is that the fitis not to the real data but to the noise. So at a givenpoint in time we have more confidence in the biologicalsignificance in those transcription patterns with largerweights. Even so, we will see that the fourth transcription pattern is very important at early times.
mentation of co-clusteringmethods to detect similar expression patterns, e.g., [10]. The maximal entropy method has been used to identify association of genes [11,12]. We too assume that the entropy depends o
Amendments to the Louisiana Constitution of 1974 Article I Article II Article III Article IV Article V Article VI Article VII Article VIII Article IX Article X Article XI Article XII Article XIII Article XIV Article I: Declaration of Rights Election Ballot # Author Bill/Act # Amendment Sec. Votes for % For Votes Against %
COUNTY Archery Season Firearms Season Muzzleloader Season Lands Open Sept. 13 Sept.20 Sept. 27 Oct. 4 Oct. 11 Oct. 18 Oct. 25 Nov. 1 Nov. 8 Nov. 15 Nov. 22 Jan. 3 Jan. 10 Jan. 17 Jan. 24 Nov. 15 (jJr. Hunt) Nov. 29 Dec. 6 Jan. 10 Dec. 20 Dec. 27 ALLEGANY Open Open Open Open Open Open Open Open Open Open Open Open Open Open Open Open Open Open .
Some Applications of the Bounded Convergence Theorem for an Introductory Course in Analysis JONATHAN W. LEWIN Kennesaw College, Marietta, GA 30061 The Arzela bounded convergence theorem is the special case of the Lebesgue dominated convergence theorem in which the functions are assumed to be Riemann integrable. THE BOUNDED CONVERGENCE THEOREM.
In Unconditional Convergence (2011), Dani Rodrik documented that manufacturing industries exhibit unconditional convergence in labor productivity. We provide a novel semi-parametric specification for convergence equations and show that the pace of convergence varies systematically with country-specific characteristics.
Of the three closely associated convergences—technological convergence, media convergence, and network convergence—consumers most often directly engage with technological convergence. Technological convergent devices share three key characteristics. First, converged devices can execute multiple functions to serve blended purpose.
Keywords: Open access, open educational resources, open education, open and distance learning, open access publishing and licensing, digital scholarship 1. Introducing Open Access and our investigation The movement of Open Access is attempting to reach a global audience of students and staff on campus and in open and distance learning environments.
Network Blue Open Access POS Blue Open Access POS Blue Open Access POS Blue Open Access POS Blue Open Access POS Blue Open Access POS Blue Open Access POS Contract code 3UWH 3UWF 3UWD 3UWB 3UW9 3UW7 3UW5 Deductible1 (individual/family) 1,500/ 3,000 1,750/ 3,500 2,000/ 4,000 2,250/ 4,500 2,500/ 5,000 2,750/ 5,500 3,000/ 6,000
for problem solving in complex systems . single-domain methods. Further defining S&T convergence “Convergence of Knowledge, Technology and Society”, 2013 et al (Refs 6- 10) Convergence approach includes: -Deep integration of knowledge, tools and modes of thinking driven by unifying concepts & a common goal-To form a new framework, paradigm or ecosystem-From where emerge novel pathways .