Eugene Shakhnovich Chemistry And Chemical Biology Harvard .
Climbing the scale ladder in biologyLecture 3: Biophysical walks on fitnesslandscapes of viruses and BacteriaEugene ShakhnovichChemistry and Chemical BiologyHarvard UniversityENS mini seriesMay 27, 2015
Length scales in biology and evolutionProbability of Fixation:PopulationPfix 1- exp(2s)1- exp(N e s)Part 2:NorvirusescapeFitness Effect:OrganismPart 1:E. GCTAAfitnessmutant - fitnesswildtypes otein-protein interactionProtein-DNA interaction
Exploring Fitness Landscape of E.ColiBottom UpA Problem:Analysis of existing sequences gives a biased view offitness landscape because mutations that we observehave been selected.Can be interpreted only using an evolutionary modelA Solution:Introduce controllable genetic variation and evaluateits fitness consequences without interveningselection.
Biophysics is a stepping stone to linksequences to phenotype
The experimental setupin vitroin silico12Identify mutations withdesired effect on stabilityand activityGenerate constructsfor recombinantexpressionDG, Tm, kcat/KmMeasure molecularproperties of thepurified proteinsin vivoIntroduce mutation(s) on the chromosome bysite-directed mutagenesisendogenous PcmR5folAkanRMeasure DFEcompetition with wt strainGenotype-phenotype linkage43pETBershtein S .&Shakhnovich EI. PNAS 2012
Determination of the bacterial growthparameters: ‘lag’ l and growth rate m:𝑦 𝐴 exp exp0.8This is a Gompertz’s equation usuallyused for fitting bacterial growth curvesin ln(OD/OD0) scale.Here,A height of the plotmm the growth rate (slope at themidpoint of the transition)l lag time (time where the tangent atthe inflexion point has y 0)OD6000.60.40.20.0lag0𝜇𝑚 𝑒𝐴 𝜆 𝑡 1120240360Time (min)480
Biophysics is a stepping stone to linksequences to phenotype
Theory: Evolution of Stability as a Random Walk(Biased Diffusional Motion) on the ‘’Fermi-Dirac’’Fitness LandscapeASSUMPTION: Fitness # of folded proteinsMutational wind - stronger at higher mFolding Free EnergyDeath cliffselection stronger with N
DHFR Is An Essential Core Metabolism Enzyme;Expressed in Low copy number of 100/cell
Adenylate kinase- a high copy # stand-alone enzyme.Expressed at 10,000 copies per cellNMP 214 amino acids inE. coli variant Three structuraldomains:LIDLID (ATP binding): 118-160NMP (AMP binding): 30-67CORE: 1-29; 68-117; 161-214 Large conformationaldynamics of LID andNMP domainCORE𝐴𝑇𝑃 𝐴𝑀𝑃𝑀𝑔 2𝐴𝐷𝑃
Selected mutations based on sequence analysisG080A, G080NL082F, L082I, L082VL083A, L083E, L083F, L083I, L083TMuts: did not occur in MSAMuts: with higher conservationMuts: with varying degrees ofconservationA093F, A093I, A093L, A093V, A093YM096L, M096NV106A, V106H, V106L, V106N, V106WY182A, Y182F, Y182VL209A,L209F, L209I, L209SL213A, L213FA total of 31 mutants at 9positions were selected
Location of the selected mutationsAll mutations are in the CORE domain and are buried
Thermal denaturation using DSCBlack trace is for WT
Chemical denaturation by CD using ureaBlack trace is for WTCD signal (mdeg)0-5-10L83E-15-20-2501234Urea (M)567
2.453.2-4-8-12-16-20-2.0-1.5-1.0-0.50.0Cm(M)Almost all the mutants aredestabilizing, as expectedCm and Tm correlate very well,indicating similar unfoldingmechanism (2 state unfolding)* Fit not reliable0.5
2Ifluorescence (au)ANS fluorescence (au)Aggregation andmolten globuleWTL83EV106H400440480520560600640wavelength (nm)ProteoStatANSIncreasing stability
16300122258150475KM ( M)kcat (min -1)Activities of all mutants have been measuredA positive correlation betweenstability and activity00-16-12-8-4Tm (oC)0-16-12-8-40Tm (oC)17
Fitness landscape for AdK projected onthe stability axis: a perfect cliff:Growth 0444852564044485256Tm (ºC)18
Fitness landscape for Adk fits roughly the‘’Fermi-Dirac’’ 68 0246G (kcal/mol)8 02468
Quite surprisingly,evolution optimizes lag time as well!:Wt stays just above the cliff:-20Lagapp02040601001200.00 0.04 0.080.200.400.60Catalytic ContingencyCatalytic contingency Protein Abundance*kcat/KM20
DHFR: (100 molecules per cell)26 Point mutations in DHFR (Bershtein et al PNAS’12),% conservationE.coli’ DHFR
Thermodynamic, structural, and catalytic propertiesof the in vitro purified DHFR rshtein S .&Shakhnovich EI. PNAS 2012
30oC1.510.5000.511.52oFitness (predicted), 30 C237oCFitness (measured), 42oC2Fitness (measured), 37oCFitness (measured), 30oCFitness is at the plateau at low T butinversely correlated with protein stability at elevated T1.510.5000.511.52oFitness (predicted), 37 C2.542oC21.510.5000.511.52oFitness (predicted), 42 CBershtein S .&Shakhnovich EI. PNAS 2012
Why?Shimon on odd daysShimon on even daysme
Fitness prediction based on protein thermodynamicsassumes:1. Equilibrium2. No flux of matter or energy
Free EnergyDestabilization can promoteoligomerization and/or aggregationDGmut DG Destabilization(mutation, heat)DGdimerizationmonomers(Partially) unfoldedmonomersoligomerAssociation pathwayLiu Y & Eisenberg D. Protein Science 2002
Cross-linking experiment shows in vitrooligomerization of high fitness and aggregation of low fitnessmutant DHFRs at high temperature(W133V)(I155A)DHFR mutants can be roughly dividedinto two groups based on their solubilityat 42C
Soluble oligomerization of the DHFR mutantscontributes to improved fitness at high temperatureBershtein S .&Shakhnovich EI. PNAS 2012
Why are these 5 mutant strains lethal? probably they fail to fold properly.(and fail to oligomerize as well)Finkelstein,Shakhnovich,Biopolymers,1989A fluorescent hydrophobic dye ANS is an excellent probe for MG state - it fluorsescesProteins like materials can be in three states: crystal-like (Native), liquid-like (Moltenupon penetration into MG but does not interact with N (too rigid) or RC (noGlobule) and gas-like(Randomhydrophobicclustersto bind) Coil)29
DHFR of low fitness mutants are Molten Globules:Evidence from ANS fluorescence in vitroLethal Mutants
Mutants which exhibit high Molten Globule Content do not growwell at 42C but are rescued by GroEL overexpression or by theknockout of an MG-sensitive protease Lon.Lon and GroEL compete for Molten Globule DHFR in thecytoplasm hence their symmetric action.Mol Cell 201331
GroEL overexpression affects fitness of each mutant inthe same way as Lon knockoutMol Cell in press32
How about Adk?Chaperone over expression does not rescue AdKmutants, unlike DHFR
Why does GroEL act onfitness same as Lon?Image credit:Ruah Edelstein34
Active cytoplasm environment vs protein solution:key differencesProtein solution:mutations affectthe distributionbetween foldedand unfoldedStates accordingtoBoltzmann law35Active environment:Mutations affect thebalance betweenproduction anddegradation, i.e.of proteinsin the cell
The ‘’Corral Reef’’ model:A simplest kinetic model of active cellular milieu,no aggregationF – folded proteinIB – intermediate bound to GroELIU – free (unbound) intermediateC – production of protein molecules at steady-statek0 – degradation of F (constant and unrelated to Lon protease)
Soluble abundance of DHFR mutants isproportional to signature ANS fluorescence,as predicted by the modelB1.5Soluble abundance at 30 oCSoluble abundance at 30 oCA10.500510152025o30ANS Fluorescence, 30 CFig. 5Mol Cell 2013, v.49, p1331.510.5000.51/AN
At steady-state:Key prediction of the dynamic theory:Lon knockout is an equalizer:Dlon growth rates do not depend on stability!CG Gmaxas k Lon 0C F0 k0Mol Cell 2013, v.49, p.133
The genetic proteostatic network of folA withGroEL/ES and Lon:Full stochastic simulationdegradedkr0[DNA]d F0kP0[mRNA][FolA F]kF0 - Idr0degraded[FolA I]kIeff- F kI0- F kIGroEL/ES [GroEL / ES]- Fdegradedd eff dI0 dILon [Lon]Gillespie algorithm, 3000 trajectories simulatedMuyoung Heo
Lon knockout in simulations indeedequalized DHFR abundance for allmutants
Lon knockout indeed makes all the ‘’phylogeny’’DHFR strains grow with (approximately) same rates.41Mol Cell, 2013, v.49, p.133
Why?RsProtein synthesisDegradation by LonDegradation by other proteasesWhen Lon is knocked out the F-I interconversion becomesThe amount of F in the cytoplasm is determined by the balancebetween production of F and its degradation42
Biophysics is a stepping stone to linksequences to phenotype
Projecting bacterial fitness to biophysicalcoordinatesE. coliDihydrofolate reductaseDHFR (folA gene)Kacser & Burns Genetics 1981Flux dynamics theory:æ kcat öæ 1 öAç çbDG è K m øè 1 e øfitness º flux µæ kcat öæ 1 öconstant A ç ç bDG øè K m øè 1 e “Michaelis-Menten-like’’dependence on abundance Aand activity Boltzmann-like dependence onstability
Orthologous DHFRs from diverse mesophiles20Frequency151050102030 40 50 60 70 80 90 100SequenceSequenceidentityIdentitywith respect toE. coli DHFR, (%)(with Shimon Bershtein)
Orthologous replacements on E. coli chromosomeallow to explore broadly the biophysics-fitness mapOrthologous replacement in the chromosomewhile preserving the endogenous promoter6cmRMeasure fitnessfolA OrthologkanRln(OD/OD0)5endogenous P43210050 100 150 200 250 300 350 400Time (min)E. coliGrowth rate:(slope of exponential phase)42 orthologous DHFRs have been purified and theirbiophysical properties in solution/cell(stability, activity, abundance in cytoplasm etc) have been determined.*That allows us to map biophysics to fitness in a broad range of properties:Large steps on FL.*6 strains with unsuccessful othologous replacements.
Distribution of molecular properties and fitness effectsamong orthologous DHFR showthe major ‘’fitness barrier’’ to interspecies replacementsStability:Activity:Fitness (Growth Rates)
The cause of HGT fitness barrier is partial loss of DHFRfunction rather than toxicity of foreign DHFRNo toxicity: Adding orthologous DHFR from pBAD plasmid does not affect fitness of wt EcLack of DHFR function in the HGT strains:Adding wt DHFR from pBAD plasmidrestores fitness of HGT strains48
Propagating the bacteria under neutral evolution36 strains with DHFRchromosomal replacementsBefore evolution:E. coliLab evolution of each strain at 8 replicates: M9 glucose thiamine 37 Celsius Passage 2x per dayAfter evolution:3 weeks ( 500 generations)
Fitness effect of HGT before and after evolution does notdepend on the evolutionary distance between Ec andorthologous DHFRBlue symbols – after propagationOrange symbols – just after HGT50
Fitness Landscape for HGT:Projecting fitness into molecular properties51
Fitness Landscape for HGT:Projecting fitness into molecular properties52
Fitness Landscape for HGT:Projecting fitness into molecular properties53
Fitness Landscape for HGT:Projecting fitness into molecular propertiesParabola-like dependence oncharge of transferred DHFR-upon HGTLess so after propagation54
Next Generation Sequencing of the Evolved StrainsQ: What did evolution do to remove fitness barrier(s) to HGT?55
Next Generation Sequencing of the Evolved StrainsQ: What did evolution do to remove fitness barrier(s) to HGT?A: It inserted the IS186 186 bp mobile element upstream of lon protease and inactivated it!56
Inactivation of lon protease in the evolved strains:Direct Evidence from Western Blot with anti-lon AbApparently lon distinguishes between ‘’self’’ and ‘’non-self’’ proteins.The mechanism of recognition is unclear57
Evolution dramatically increased DHFRabundance in cytoplasm58
Part II:Predicting evolution:Biophysical Fitness Landscape ofViral Escape from Antibodies59
Norovirus( )ss-RNA, 7.5 kbpDonaldson et al. Nat Rev Microbiol 2010.The Guardian“Most common cause of acute gastroenteritis in the United States.“Most common cause of foodborne-disease outbreaks in the United States.“Causes 20 million illnesses and contributes to 70,000 hospitalizations”-Center for Disease Control and Prevention, US
Viral evolution in the labSerial passaging:Total volume: 2 mLNumber of cells:8 x106 cells/mLNumber of viral particles: 106 pfu/mL 108 viral particles/mLBurst size: 104 virions/infected cellMutation Rate: 1 mutation/genome/generationNext-gen sequencing (Illumina):59186293250 bp250 bp
Viral evolution in the labAllele Frequency10.80.6Wildtype0.40.2Other variants00123Log (Allele Frequency)GenerationViral population ispolymorphic because ofhigh mutation rate.0123GenerationSerohijos & Rotem In preparation.
Estimating viral fitness from deep sequencingScenarios:Allele Frequency1Fitnessmutant WTFitnessmutant WTMutation was lethal0.80.6Wildtype0.40.2Other variants00123Gen iGen iGen (i 1)Gen (i 1)Gen iGen (i 1)GenerationLog (Allele Frequency)Formalism:niB niA (1 ri B)ri replication probability of clone i (fitness)B burst size (number of progenies per replication)fi B fi A (1 ri B)All clonesåfkA (1 rk B)k 10123riFitness of variant iGenerationSerohijos Rotem, ES In preparation
Estimating the biophysical effects of mutationsEffect on folding stability:CapsidP-domainXDDG folding º DGmutant - DGwildtype» Emutant - EwildtypePhysical force field:Yin et al. Nat. Methods 2007
Evolution under explicit selection pressureSerial passaging:Total volume: 2 mL AntibodyNumber of cells:8 x106 cells/mLNumber of viral particles: 106 pfu/mL 108 viral particles/mLBurst size: 104 virions/infected cellAntibody concentration: 9 nM ( 1300 ng/mL) 150 antibody molecules per virionMutation Rate: 1 mutation/genome/generation
Evolution under antibody pressure1WildtypeAllele Frequency0.80.6E296K-T301I-D385G0.40.28169 Other mutants001234PassageCapsid P-domainE296KT301ID385GAb Fab
Projecting viral fitness to capsid folding stabilityæ fraction genotypes with r 0 öPinfectivity ç all genotypesèøDataTheoryAssuming:Infectivity µ( Fraction of folded capsid proteins)Theoretical Fit:æö1Pinfect b0 ç è 1 e b (DGWT DDG) øThere is no fitness penalty for stabilizing mutations.Dependence of fitness on folding stability obeys simple thermodynamics.
Projecting viral fitness to the affinity to antibodyæ fraction genotypes with r 0 öPinfectivity ç all genotypesèøDataTheoryAssuming:æ Fraction of capsid proteins öInfectivity µ ç è unsequestered by AbøTheoretical Fit:Thermodynamics of protein-protein interaction:A B « A·Bæ K [ Ab] ö Fraction unbound çç d1 KAb[]èødFree unboundæ K [ Ab] ö Pinfectivity b0 çç dè 1 K d [ Ab] øKd /[Ab]
Dissect theevolutionarypathwaysProtein biophysical properties are principal coordinates of the fitness landscape.
Experimental measurements of folding stabilityand affinity to antibodyBSAThermal fluorescence:Niesen et al. Nature Protocols 2007.WTkDaSurface Plasmon Resonance:
Experimental measurements of folding stabilityand affinity to antibody1.00E-04Escapee during bulk expansionafter prior droplet evolution, Expt #2Escapee during bulk expansionafter prior droplet evolution, Expt #1Dissocia on Constant, rop 1Ibulk .537.037.538.038.5Mel ng Temperature, Tm39.039.540.0
Adaptation along the biophysical coordinates1Allele 4PassageSerohijos & Rotem In preparation.
Viral population climbs to a fitness type
Adaptation along the biophysical coordinates1Allele assage34Why does Tm follow Kd?Serohijos & Rotem In preparation.
Relationship between stability andbinding affinity among random mutationsRandom single substitutions from WT(Computational Estimate)ΔΔGbinding (kcal/mol)10864Virus escapesantibody20y -0.5x 0.8R² 0.37-2-4-6-6-4-20246810ΔΔGfolding (kcal/mol)Less likely escape routeMost likely escape routeKdKdTmLeast likely escape routeKdTmTm
Forward Simulation on the biophysicalfitness landscapeDGwt -4 kcal/mol
Viral population climbs a local fitness peakE296K-T301I-D365G1 lab evolutionexpt.Can we bypass this localfitness peak?E296K-T301I-D365GE296K-T301I3 lab tion of other biophysical propertiesTwoconformersAb bindingregions(Tom Smith, U Texas)Mutations in other regions of the genome
Exploration of a rugged fitness landscape isAnalogyBetweenEvolutionarycontrolledby populationsizeBiologyand Statistical MechanicsEvolutionary Biology(Wright’s Shifting Balance Theory)Sample largersequence spaceSmall N(Weak or No selection)ANRV328-ES38-02ARIStatistical Mechanics(Simulated Annealing)High TPhase 224 September 2007Sample largerconfiguration space8:50Phase 3Phase 1Ecol. Evol. Syst. 2007.38:27-52. Downloaded from www.annualreviews.orgby Harvard University on 07/21/13. For personal use only.Large N(Strong selection)Low TFitness landscapePopulation 1Population 2Population 3Free Energy landscapeFitnessFitnessFitnessEvolutionary “simulated annealing” by cycling through
Rational control of population size:Viral evolution in droplet emulsionsReinjected nCellsWasteAssaf Rotem & Dave Weitz, SEAS, Harvard
Evolutionary “Annealing” #1Droplet evolutionBulk (3 passages)Allele Frequency10.80.60.40.20012345Passage99
Evolutionary “Annealing” #2Droplet evolutionBulk (3 passages)Allele Frequency10.80.60.40.200123Passage459
Evolutionary “Annealing” #2Droplet evolutionBulk (3 passages)Allele Frequency10.80.60.40.200123459PassageAn unlikely path withoutevolutionary “annealing”.
ection of viral evolution can becontrolled by modulating population size
Validating the Escape Variants Using ReverseEngineered Mutants1.00E 091.00E 081.00E 071.00E 061.00E 051.00E 041.00E 031.00E 021.00E 011.00E 000 mAb60 ng/ml mAb200 ng/ml mAb600 ng/ml mAbMutant Sensitivity to A6.2.1(% Infectivity)% Infectivitypfu/mlMutant Sensitivity to 0200600Joshua Wolf (Johns Hopkins APL)
AcknowledgementsHarvard University (ES lab, Chemistry)Harvard University (Applied Physics) Adrian Serohijos David Weitz Shimon Bershtein Assaf Rotem Bharat Adkar Sanchari Bhattacharyya Andrew Feldman Michael Manhart Jeffrey Lin Ryan Lee Tom Mehoke Michael Zimmet Audrey Fis
Fitness (predicted), 30oC 0 0.5 1 1.5 2 2.5 0 0.5 1 1.5 2 Fitness (predicted), 42oC 2 o C 0 0.5 1 1.5 2 0 0.5 1 1.5 2 Fitness (predicted), 37oC 7 o C Fitness is at the plateau at low T but inversely correlated with protein stability at elevated T B
North Coast Chorale Astoria 7,517 North Coast Symphonic Band (NCSB) Astoria 2,849 . Emerald Empire Art Association (EEAA) Springfield 7,326 . 7,326 Eugene Ballet Company Eugene 264,699 Eugene Concert Choir Eugene 32,609 EUGENE GAY MEN'S CHORUS Eugene 3,566. Eugene Gleemen Eugene 5,826 Eugene Opera Eugene 173,578 .
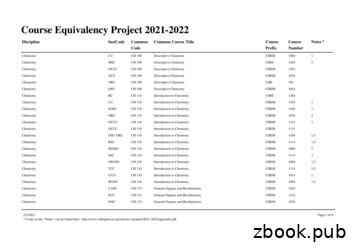
Chemistry ORU CH 210 Organic Chemistry I CHE 211 1,3 Chemistry OSU-OKC CH 210 Organic Chemistry I CHEM 2055 1,3,5 Chemistry OU CH 210 Organic Chemistry I CHEM 3064 1 Chemistry RCC CH 210 Organic Chemistry I CHEM 2115 1,3,5 Chemistry RSC CH 210 Organic Chemistry I CHEM 2103 1,3 Chemistry RSC CH 210 Organic Chemistry I CHEM 2112 1,3
Physical chemistry: Equilibria Physical chemistry: Reaction kinetics Inorganic chemistry: The Periodic Table: chemical periodicity Inorganic chemistry: Group 2 Inorganic chemistry: Group 17 Inorganic chemistry: An introduction to the chemistry of transition elements Inorganic chemistry: Nitrogen and sulfur Organic chemistry: Introductory topics
Volunteer Experience ArtCity Eugene, event coordinator and artist, 2015 - present, Eugene, OR Maude Kerns Art Center, Board of Directors, 2017 – present, Eugene, OR Eugene Mission, Kitchen staff, winter 2018 Singing Creek Educational Center, Pioneer Guide, 2016, Eugene, OR Richard App Gallery, framing/gallery assistant, 2012-2013, Grand Rapids, MI
ADVANCED DIPLOMA Diploma in Chemistry 60% in Analytical Chemistry 3 Theory & Practical, Chemical Quality Assurance, Mathematics 2 Chemical Industrial 1 or S5 Subjects and Chemistry project II. Semester 1 Analytical Chemistry IV Physical Chemistry IV Research Methodology in Chemistry Semester 2 Inorganic Chemistry IV Organic Chemistry IV .
Chemical Formulas and Equations continued How Are Chemical Formulas Used to Write Chemical Equations? Scientists use chemical equations to describe reac-tions. A chemical equation uses chemical symbols and formulas as a short way to show what happens in a chemical reaction. A chemical equation shows that atoms are only rearranged in a chemical .
Levenspiel (2004, p. iii) has given a concise and apt description of chemical reaction engineering (CRE): Chemical reaction engineering is that engineering activity concerned with the ex-ploitation of chemical reactions on a commercial scale. Its goal is the successful design and operation of chemical reactors, and probably more than any other ac-File Size: 344KBPage Count: 56Explore further(PDF) Chemical Reaction Engineering, 3rd Edition by Octave .www.academia.edu(PDF) Elements of Chemical Reaction Engineering Fifth .www.academia.eduIntroduction to Chemical Engineering: Chemical Reaction .ethz.chFundamentals of Chemical Reactor Theory1www.seas.ucla.eduRecommended to you b
The American Board of Radiology . i The Diagnostic Radiology Milestone Project The Milestones are designed only for use in evaluation of resident physicians in the context of their participation in ACGME accredited residency or fellowship programs. The Milestones provide a framework for the assessment of the development of the resident physician in key dimensions of the elements of physician .