Fundamentals Of Protein Chemistry - Duke University
Fundamentals of Protein ChemistryAmino Acid and Peptide ChemistryTranscription and TranslationPost-Translational ModificationsClassical Analytical MethodsIn-Gel Proteolysis
Amino Acids are the Basic Structural Units of Proteins
Names, Abbreviations, and Properties of The Twenty Amino AcidsNameAlanineArginineAspartic AcidAsparagineCysteineGlutamic ptophanTyrosineValineSide Chain 16.35.6810.95.885.636.006.0
The Twenty Amino Acid Side Chains Vary SignificantlyThe twenty naturally occurringamino acids that make upproteins can be groupedaccording to many criteria,including hydrophobicity, size,aromaticity, or charge.http://www.imb-jena.de/image library/GENERAL/aa/chemprop.jpg
Amino Acids are Linked by Amide Bonds to Form Peptide Chains
Acid-Base Chemistry in Protein CharacterizationThe net charge of a peptide or protein at any pH depends on thecombined pK values for its amino acids and terminal groups.pH pK log [A-]/[HA]pK of alpha-COOH groups:pK of alpha-NH2 groups:pK of ionizable side chains:1.8 - 2.49.0 - 10.83.9 - 12.5The isoelectric point is the pH at which there is no net charge.It is important to remember how protein and peptide pK valuesaffect chemistry and separations:Chemical Modification (e.g. Reduction / Alkylation)Proteolysis (e.g. specificity of Glu-C)Chromatography (e.g. Ion Exchange)2D Gel Separations (Isoelectric Focusing)Ionization for Mass Spectrometry (e.g. MALDI-TOFMS)
Peptide Chains are Linked by Disulfide Bonds Between Cysteines
Protein Synthesis:Transcription & TranslationMessenger RNA is synthesizedin the nucleus and exits througha nuclear pore.Ribosomes assemble along themRNA and translate it into apeptide sequence.The growing peptide may bedirected into the lumen of theendoplasmic reticulum (ER) forprocessing.T. Zamis, U. Wisc., Stephens mistry.html
Translation: Amino Acid sTGTTGCTrpTGGArgCGTCGCCGACGGAGAAGGGlyGGTGGCGGAGGG
Protein Synthesis: TranslationHistology by Bergman, R.A., Afifi A.K. and Heidger, P.M. Saunders Publishing,Philadelphia, Pa, 1995Molecular Biology of the Cell by Bruce Alberts, Dennis Bray, Julian Lewis,Martin Raff, Keith Roberts, and James D. Watson, Garland Publishing, NY 1994
Protein Synthesis: TranslationAs a peptide grows into the ER, it is folded and modified.The ER has multiple compartments in which specific bindingproteins and enzymes act on the new protein molecule.Histology by Bergman, R.A., Afifi A.K. and Heidger, P.M. Saunders Publishing,Philadelphia, Pa, 1995A Textbook of Histology by D Fawcett.Chapman and Hall, N.Y. 1994
Protein Folding, Modification, and TransportA newly synthesized protein can be transported from the ERto the Golgi Apparatus, another complex series ofcompartments in which modifications are made. The Golgiis important for determining the disposition of proteins.A Textbook of Histology by D Fawcett. Chapman and Hall, N.Y. 1994Molecular Biology of the Cell by Bruce Alberts, Dennis Bray,Julian Lewis, Martin Raff, Keith Roberts, and James D. Watson,Garland Publishing, NY 1994
Protein Post-Translational ModificationProtein modifications performed by “extra-translational” processes.Cannot be definitively predicted from DNA sequenceCan involve very complex systems of enzymesIn some cases, “consensus” sites of modification can be identifiedUbiquitous in eukaryotesFrequently critical for:initiation or modulation of biological activitytransport, secretion
Protein Post-Translational ModificationProteolytic CleavageN-term Met of all proteins removed by aminopeptidasesN-terminal Acylationformyl, acetyl, myristyl, etc. by acyltransferasesGlycosylation Asn, Ser, and ThrSulfation TyrPhosphorylationSer, Thr, and TyrCarboxyl Terminal AmidationHydroxylation Pro, Lys, AspN-Methylation Lys, Arg, His, GlnCarboxylationGlu, AspModifications introduced by usMet [O], ees/deltamass/deltamass.htmlExhaustive list maintained by Ken Mitchelhill
Protein MutationsSingle nucleotide changes can result in amino acid substitutionsSingle amino acid substitutions can result in alterations in:Protein 3D structureMolecular WeightIsoelectric PointProteolysis by specific enzymesMolecular weights of proteolytic fragmentsProtein Prospector, UCSF MS Facilityhttp://prospector.ucsf.eduTable of Mass Shifts due to Single AA sc/mutation.htm
Robust Analytical Methods for Protein CharacterizationAmino Acid AnalysisAcid Hydrolysis followed by derivatization and HPLCDetermines the precise molar ratios of amino acids presentCan also be used to accurately determine concentrationAsp/Asn and Glu/Gln are not distinguishedCysteine and Tryptophan are problematic in some methodsAmino-Terminal Sequencing by Edman DegradationVery sensitiveStandard method- still best approach to NH2-terminusVery steep learning curve to do it wellBlocked proteins cannot be analyzedMixtures are challengingPeptides with long repeats are problematicPTMs are often missed but can be dealt withOften not competitive with MS for internal sequence
Robust Analytical Methods for Protein CharacterizationPolyacrylamide Gel ElectrophoresisA crude measure of molecular weight and purityAnalytical or preparative separationsCoupled with Blotting- sensitive & selective detectionIsoelectric FocusingAnalytical or preparative separationsUsed for mapping disease markers (e.g. CGDs)Variety of pH gradientsAutomated, high throughput instrumentsTwo Dimensional IEF – PAGEOrthogonal separations- large separation spaceDetection of small changes in complex samplesSeparation of post-translationally modified proteinsDynamic Range problems due to sample loading capacity
Isoelectric FocusingIn a pH gradient, under an electric field, a protein will move to the position in thegradient where its net charge is zero.An immobilized pH gradient is created in a polyacrylamide gel strip by incorporatinga gradient of acidic and basic buffering groups when the gel is cast.Proteins are denatured, reduced, and alkylated, and loaded in a visible dye.The sample is soaked into the gel along its entire length before the field is applied.Resolution is determined by the slope of the pH gradient and the field strength.Immobilized pHgradient gel stripsMany can be run inparallel for greaterreproducibility
Polyacrylamide Gel ElectrophoresisPolyacrylamide gels may be have manydifferent constant or gradient compositions.Proteins may be run directly, or aredenatured, reduced, and alkylated.Samples, with a visible dye added, are loadedin wells cut into the top of the gel.Loading capacity depends on gel size andthickness.In 2D IEF/PAGE, the gel strip from IEF isloaded into a single large well.Molecular weight standards are often run tocalibrate the gel.After separation, the gel is removed from therig and stained, or bands are blotted onto amembrane.
Chemical Methods for Protein CharacterizationDenaturation- dissociates and unfolds proteinsChaotropes: Urea, GuanidineDisulfide Bond CleavageReducing Agents: Dithiothreitol, beta-mercaptoethanolCysteine Alkylation- prevents reoxidation to form disulfidesAlkylating Agents: Iodoacetamide, Vinylpyridine
Chemical Methods for Protein Characterization: ProteolysisChemical Methods:Acid Hydrolysis, various [H ], time, tempCyanogen Bromide cleavage, C-term to Methionine gives a homoserine lactone at MetEnzymes:TrypsinChymotrypsinS. aureus V8 proteaseS. aureus V8 proteaseAchromobacter protease (Lys-C)Arg-CAsp-NThermolysinC-term to Lys, ArgC-term to Y, F, W, H, LC-term to GluC-term to Glu, AspC-term to LysC-term to ArgN-term to AspN-term to L, I, M, F, WpH 8.5pH 8.5pH 8pH 5pH 8pH 8pH 8pH 8.5Specificity of Proteolysis is a very powerful weaponSpecificity of these methods is variable: some excellent, some almost noneLadder Sequencing, sometimes done using MALDI-TOF/MSManual Edman Degradation, Carboxypeptidase Y digestion, Acid Hydrolysis
Chemical Methods for Protein Characterization: ABasic Protocol for Denaturation & Proteolysis1. TCA Precipitation:(if [protein] is 0.05 mg/mL)Chill protein in a microcentrifuge tube to 0 CAdd 1/9 volume of cold 100% w/v TCA. VortexIncubate at 0 C for 30-60 minSpin down in microcentrifuge. Remove supernatantWash pellet 3x w/ 200 μL cold acetone (do not vortex)Air dry pellet 30 min2. Redissolving the sample in a chaotrope:Redissolve protein in 50 μL of fresh 8 M urea/0.4 M Amm. Bicarb.For efficient digestion, [protein] of 0.025 μg/mL is required.(Final volume 4x the volume of urea added)-adjust as necessary3. Reduction and alkylation of cysteines:Add 5 μL (or 1/10 volume) of 45 mM DTTIncubate at 50 C for 15 min. Cool to room temperatureAdd 5 μL of 100 mM iodoacetamideIncubate in the dark at room temperature for 15 minBarbara Merrill, Bayer Corp.
Chemical Methods for Protein Characterization: ABasic Protocol for Denaturation & Proteolysis4. Trypsin digestion:Add 140 μL of ddH2O, vortex. Check that pH is between 7.5 and 8.5.Trypsin added should be a 1:25 weight:weight ratio of protease to sampleConcentration of trypsin should be such that 1 to 5 μL is added to sampleIncubate at 37 C for 24 h. Stop digest by freezingNotes:Trypsin used should be treated with TLCK to inhibit residual chymotrypsin.Trypsin is made up at 1 or 5 mg/mL in 1 mM HCl.Aliquots can be stored frozen for up to 3 mos (use once and discard).This protocol can be used for chymotrypsin or Achromobacter protease (Lys-C)Final [urea] of 1 M is suitable for Staph. Aureus V-8 protease (Glu-C) or Asp-N proteaseIn the case of Asp-N a 1:50 to 1:100 w:w ratio should be used; digest for 16 hBarbara Merrill, Bayer Corp.
Chemical Methods for Protein Characterization:In-Gel ProteolysisWhy In-Gel Digestion Works:1. The gel piece behaves like a sponge. It shrinks and swellsin response to addition of aqueous or organic solvent.A gel piece shrunk with organic solvent will suck in anaqueous buffer containing reagents, thus bringing them“inside” the matrix to access the protein.2. The intact protein is trapped in the gel, so many chemicalsteps can be performed without significant loss.3. Many of the peptides resulting from proteolysis within thegel freely diffuse out of the matrix.
Chemical Methods for Protein Characterization:In-Gel Proteolysis1. Destaining the Gel Band:Mince gel spot or band with razor blade (cut into 1 mm cubes).Wash gel pieces with 50% acetonitrile in 50 mM Ammonium bicarbonate pH 8.00.5 – 1.0 ml depending on the volume of gel pieces; wash 20 min. on rotator.If volume of pieces is large (greater than 500 ul) do 2 washes at 20 min each.Shrink pieces with neat acetonitrile.Dry in speed vac.Mary B. Moyer, GlaxoSmithKline, after “Femmtomole sequencing of proteins from polyacrylamide gels by nano-electrospray massspectrometry”, M. Wilm, A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, M. Mann; Nature, 1996, 466-469.
Chemical Methods for Protein Characterization:In-Gel Proteolysis2. Reduction and alkylation:Buffer:6 M guanidine HCl0.5 M ammo. Bicarb. PH 8.05 mM EDTATo 200 ul of above buffer, add 2 ul of 2 M DTT & 0.5 ul 2- or 4-vinyl pyridineAdd just enough to cover the re-swollen gel piecesIncubate 15 min at 37C or 30 min RTRemove excess reduction and alkylation mix; wash gel pieces 2 times with 1 mlaliquots of 50 mM ammonium bicarbonate pH 8.0.Shrink with neat acetonitrile. Swell in 50 mM AmBic pH 8.0.Shrink with neat acetonitrile. Swell in 50 mM AmBic pH 8.0.Shrink with neat acetonitrile. Dry in speedvac.Mary B. Moyer, GlaxoSmithKline, after “Femmtomole sequencing of proteins from polyacrylamide gels by nano-electrospray massspectrometry”, M. Wilm, A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, M. Mann; Nature, 1996, 466-469.
Chemical Methods for Protein Characterization:In-Gel Proteolysis3. Proteolysis:Swell in 50 mM AmBic pH 8.0 containing 12.5 ng/ul trypsin.Add enough trypsin solution to cover fully swollen gel piecesDigest for 16 – 18 hours.4. Analysis:For MALDI-TOF/MS: an aliquot may be taken from the supernatant around the gelpiecesFor LC/MS: all the volume plus an additional extraction of the pieces in 50 mMAmBic pH 8.0 may be combinedMary B. Moyer, GlaxoSmithKline, after “Femmtomole sequencing of proteins from polyacrylamide gels by nano-electrospray massspectrometry”, M. Wilm, A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, M. Mann; Nature, 1996, 466-469.
Combining Analytical Methods for Protein nal ModificationsFull coverage almost always requires multiple digests.Identification essentially never does.
a gradient of acidic and basic buffering groups when the gel is cast. Proteins are denatured, reduced, and alkylated, and loaded in a visible dye. The sample is soaked into the gel along its entire length before the field is applied.
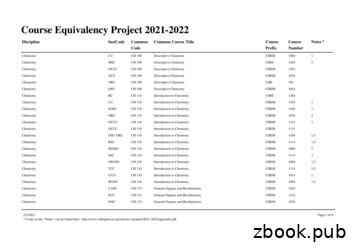
Chemistry ORU CH 210 Organic Chemistry I CHE 211 1,3 Chemistry OSU-OKC CH 210 Organic Chemistry I CHEM 2055 1,3,5 Chemistry OU CH 210 Organic Chemistry I CHEM 3064 1 Chemistry RCC CH 210 Organic Chemistry I CHEM 2115 1,3,5 Chemistry RSC CH 210 Organic Chemistry I CHEM 2103 1,3 Chemistry RSC CH 210 Organic Chemistry I CHEM 2112 1,3
MY.DUKE.EDU/STUDENTS- Personal info & important links Navigate Campus CALENDAR.DUKE.EDU-University events calendar STUDENTAFFAIRS.DUKE.EDU- Student services, student groups, cultural centers DUKELIST.DUKE.EDU- Duke’s Free Classifieds Marketplace Stay Safe EMERGENCY.DUKE.EDU-
„Doris Duke of the illuminati Duke family was an heiress (at 12 years old) to the large tobacco fortune of the Duke family. She was the only child of American tobacco Co. founder James Buchanan Duke. Doris Duke, herself a member of the illuminati. Doris Duke had 5 houses (which have served as sites for illuminati rituals) – one in Beverly
The Duke MBA—Daytime Academic Calendar 2015-16 9 Preface 10 General Information 11 Duke University 11 Resources of the University 13 Technology at Fuqua 14 Programs of Study 15 The Duke MBA—Daytime 15 Concurrent Degree Programs 17 The Duke MBA—Weekend Executive 18 The Duke MBA—Global Executive 18 The Duke MBA—Cross Continent 19
protein:ligand, K eq [protein:ligand] [protein][ligand] (1) can be restated as, K eq 1 [ligand] p 1 p 0 (2) where p 0 is the fraction of free protein and p 1, the fraction of protein binding the ligand. Assuming low protein concentration, one can imagine an isolated protein in a solution of Nindistinguishable ligands. Under these premises .
Physical chemistry: Equilibria Physical chemistry: Reaction kinetics Inorganic chemistry: The Periodic Table: chemical periodicity Inorganic chemistry: Group 2 Inorganic chemistry: Group 17 Inorganic chemistry: An introduction to the chemistry of transition elements Inorganic chemistry: Nitrogen and sulfur Organic chemistry: Introductory topics
MLO Super High Protein powder, MLO Brown Rice Protein powder, MLO Milk and Egg Protein powder, MLO Vegetable Protein powder UNJURY Protein bariatric surgery patients Optimum Protein Diet Shakes Bariatric Fusion Protein Supplement Bodytech Whey Pro 24 Premier
of protein assay for research applications. Protein assays based on these methods are divided into two categories: dye binding protein assays and protein a ssays based on alkaline copper. The dye binding protein assay s are based on the binding of protein molecules to Coomassie dye under acidic conditions.