FAME And 16srDNA Sequence Analysis Of Halophilic Bacteria .
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-31531FAME and 16srDNA sequence analysis of halophilicbacteria from solar salterns of Goa: A comparative studySurve V V*, Patil M U**, Dharmadhikari S M****Department of Biotechnology, Vivekanand College**Department of Zoology, Dr.BAMU***Department of Microbiology, Institute of Science Aurangabad, Maharashtra, 431001 IndiaAbstract- Halophilic microorganisms present in thehypersaline environments of solar salterns present a potentialsource of industrial and biotechnological applications .The Rapididentification of these microorganisms which are accurate andcost effective is a necessity in applied microbiology. There havebeen several systems developed in the past few years for rapidmicrobial identification. In the present study, GC FAME( Gaschromatography- Fatty acid methyl esters) analysis and 16srDNA sequencing were compared for identification of fourhalophilic bacteria. Four halophilic bacterial isolates obtainedfrom solar saltern sediment were analyzed . The isolates wereidentified with 16srDNA rophaeus,Corynebacteriumdiphtheria and ,Idiomarina Zobellii.However in FAMEanalysis , the RTBSA 6 library did not confirm any matchesfor Idiomarina Zobellii. Comparative analysis, indicated thatthe FAME analysis results correlates to genotypic sequencingresults. However,a halophilic bacterial FAME library should beestablished to enhance the accurate , rapid and cost effectiveinvestigation of halophilic microorganisms.Index Terms- Solar salterns, Halophilic bacteria, FAME,Sequencing, BLAST, IdentificationI. INTRODUCTIONDetermining the taxonomic composition, biomass, andphysiological status of microbial assemblages is still one ofthe greatest challenges facing ,microbial ecologists. Classicalapproaches that utilize enrichment methods for the isolation ofmicroorganisms from the environment continue to providevaluable information in biochemical, taxonomic, andautoecological studies. The primary limitations to suchapproaches are those of non culturability ,and the problem ofcharacterizing and identifying statistically relevant numbers ofisolates necessary to gain insight into the population ecology andcommunity diversity of habitats(Thompson et al., 1999). Rapidand accurate identification of bacterial pathogens is afundamental goal of clinical microbiology, but one that isdifficult or impossible for many slow-growing and fastidiousorganisms(Tang, 1998).New and exciting molecular methods,using the 16S small sub-unit ribosomal nucleic acid moleculeshave added much to our knowledge of microbial diversity(Macrae A ,2000). For many years, sequencing of the 16Sribosomal RNA (rRNA) gene hasshown that sequenceidentification is useful for slow-growing, unusual, and fastidiousbacteria as well as for bacteria that are poorly differentiated byconventional methods. The technical resources necessary forsequence identification are significant. Despite the availability ofresources, sequence-based identification is still relativelyexpensive(Patel 2001).Although it is generally regarded thatroutine identification of very common species using conventionalmethodologiesare highly accurate, we now have a moreconvenient and precise mechanism for checking theseidentifications on a molecular basis. Such studies need to beperformed and published( Janda , 2007).The 16S rDNAsequence analysis is the reliable method for halophilic t advances in thebiochemistry of microorganisms revealed that analysis of cellcomponents, such as proteins and fatty acids, can be effectivelyapplied to bacterial identification, providing the basis forchemotaxonomy(Goodfellow, 1985., Komagata,1987).Fattyacids are one of the most important building blocks of cellularmaterials. In bacterial cells, fatty acids occur mainly in the cellmembranes as the acyl constituents of phospholipids. Thesefatty acids are synthesized in certain bacteria from iso, anteiso, orcyclic primer and malonyl-CoA with or without a subsequentmodification( Kaneda, 1977.,Lechevalier 1977. ,Wilkinson1988).The occurrence of branched-chain fatty acids as majorconstituents in bacteria was first reported for Bacillussubtilis(Saito, K. 1960. )These days, fatty acids in bacterial lipidsare routinely analyzed by gas-liquid chromatography. Inaddition, other methods such as nuclear magnetic resonancespectroscopy,infrared spectroscopy, mass spectrometry, and thinlayer chromatography have been used to aid the gas-liquidchromatographic identification of fatty acids ( Toshi 1991 ).Theunique pattern of fatty acids in bacteria is the basis ofidentification in FAME analysis.The MIDI (MicrobialIdentification Incorporation) Sherlock FAME analysis is alaboratory-based system that can be used on a routine basis toidentify commonly isolated bacteria from clinical andenvironmental source( Leonard et al 1995) More than 300 fattyacids and related compounds have been found in bacteria areanalyzed in the MIDI Research and Development Laboratory.Whole cell fatty acids are converted to methyl esters andanalyzed by gas chromatography. The fatty acid composition ofthe unknown is compared to a library of known organisms inorder to find the closest match. The peaks are automaticallynamed and quantitated by the system .Branched chain acidspredominate in some Gram positive bacteria, while short chainhydroxy acids often characterize the lipopolysaccharides of theGram negative bacteria(Sasser, 2001) . The MIS, as the firstautomated CFA(Cellular fatty acid ) identification system, is anwww.ijsrp.org
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-3153accurate, efficient, and relatively rapid method for theidentification of microorganisms ( Osterhout et al 1991).Theidentification of marine isolates, is a continuing challenge formarine microbiologists. The identification of marine bacteria iscommonly based on a wide range of biochemical andphysiological tests. The major difficulties found with thistraditional approach are the need for an easily cultivable strainand the time required for the preparation of cultures (AkagawaMatsushita, et al., 1992) Solar saltpans are found worldwide andare considered extreme environments with very restrictedbiology (Litchfield, 2002).It is surmised that marineenvironments like solar saltern may yield newer strains whichmay prove to be a rich resource of new metabolites.Microorganisms that thrive in these hypersaline environmentsare called halophilic microorganisms (that is, require salt fortheir viability .The domain bacteria typically contains manytypes of halophilic microorganisms that spread over a largenumber of phylogenetic subgroups and they are moderate ratherthan extreme halophiles (Oren, 2002).Sparked by theavailability of microbial diversity data in these salterns an effortis made to study the moderately halophilic bacterial in theseextreme environments. The bacteria are analyzed for theiridentity by two methods 16s rDNA sequencing and GCFAME,.The similarity between these methods is comparativelystudied.II. MATERIALS AND METHODFourhalophilic bacterial pure cultures were obtainedfrom solar saltern sediment and designated as H1, H2, H3 and H4 .The cultures were grown on halophile medium(10% salt) at28 C for 48-72 hours. The fatty acids were extracted andmethylated to form fatty acid methyl esters (FAME). TheseFAME’s were analyzed using Gas Chromatography with the helpof MIDI Sherlock software for FAME. Aerobic library (RTBSA6.0) was referred for the analysis. The analysis was performed asper (Sasser, 2001).For 16s rDNA sequence analysis thegenomic DNA was isolated. PCR amplifications of the 16SrRNA gene, from the purified genomic DNAs, were carried outusing the primer sets 27F (5 AGAGTTTGATCCTGGCTCAG3 ) and1492R (5 -GGTTACCTTGTTACGACTT-3 ). Theamplified DNA was purified and sequencing of the target genewas done using Big Dye Chemistry, and performed as per themanufacturer's protocols (Applied Biosystems ). The purifiedextension products were separated in the ABI 3730xl DNAAnalyzer by capillary electrophoresis. Sequence data analysiswas done using ChromasPro and Sequencing Analysis software.The bioinformatics analysis was performed using NCBI BLAST(http://ncbi.nlm.nih.gov/blast) identifying the microorganismusing online databases. The analysis data from the Sequencingand FAME analysis obtained was comparatively studied.III. RESULTSThe four halophilic microbial cultures obtained from salternsediment were analyzed using FAME and sequencingapproaches. On comparing FAME results with that ofsequencing, it was found that both the methods correlate to each2other in confirmation of identityof the isolates except forisolate H4. Figure 1 shows peaks corresponding to the fattyacids identified through FAME analysis of bacterial sample H1.The MIDI Sherlock microbial identification system usingRTSBA6 method identified the organism to be Corynebacteriumdiptheriae with 0.596 Similarity index(SI). The sequenceanalysis also showed similar results (Table 1). After thecompletion of BLAST analysis the organism was identified toCorynebacterium diphtheriae strain with 73% identity match.FAME analysis identified H2 as Virgibacillus Pantothenticuswith similarity index 0.698(Figure 2).Sequence analysis alliedwith this identification ,as the after BLAST analysis the isolateH2 was identified as Virgibacillus PantothenticusstrainOLO3.(Table 2).The methods also displayed parallel results forisolate H3. Wherein the H3 isolate was identified as Bacillusatrophaeus with FAME analysis (Similarity index 0.776. (Figure3).The sequencing and BLAST identified the isolated as Bacillusatrophaeus with 100% identity match. However the isolate H4,identified as Idiomarina Zobellii100 % identity match afterBLAST analysis, did not find any matching chromatogrampattern in FAME RTBSA6 6.0 aerobic library.IV. DISCUSSIONIdentification of bacteria by conventional methods usuallyrequires 48 h after a colony has been isolated. The identificationof slow-growing or biochemically inert microbes to the specieslevel is difficult and time-consuming by conventional methods.16S rRNA gene sequences frequently provide phylogeneticallyuseful information. Signature nucleotides allow classificationeven if a particular sequence has no match in the database, sinceotherwise-unrecognizable isolates can be assigned to aphylogenetic branch at the class, family, genus, or subgenuslevel. Cost is a critical issue in the evaluation of 16S rDNAsequence analysis as a diagnostic tool. However, sequencingcosts will probably continue their rapid trend downward,bringing this technology within the reach of many microbiologylaboratories (Tang, 1998). Roohi et al in 2012 studied thehalotolerant and Halophilic bacteria from salt mines of karak,Pakistan with 16S rRNA gene sequence.As the phenotypiccharacteristics alone were not enough to differentiate thebacterial isolates and could lead to identification problems. Themain reason for this is the standardization of a conventionalmethod, when it was applied to halophilic bacteria because theirgrowth characteristic highly dependent on many factors such asNaCl concentrations, temperature, pH and mediumcomposition.All the four isolates were sucessfully sequenced andidentified in the present study.The MIDI FAME systemidentified 3 out of 4 bacterial isolates to the correct specieslevel. The results were in accordance to the findings ofChookiewattana K, 2003 .Wherein 14.1% of halophilic bacterialisolates were not identified by FAME analysis, differing the100% identification by 16srDNA sequencing. Species levelidentification of microbes is possible through MIDI Sherlockmicrobial identification. Abel and colleagues first suggestedthat microorganisms could be classified by gas chromatographicanalysis (Abel et al 1963). The Sherlock Microbial IdentificationSystem is an accurate and automated gas chromatographicsystem, which identifies over 1,500 bacterial species based ontheir unique fatty acid profiles. The system can analyze over 200www.ijsrp.org
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-3153samples per day,uses a standardized sample procedure for allbacteria, and costs about 2.50 per sample in standard laboratoryconsumables (Sasser M 2001) .The MIDI’s identification waslisted with a confidence measurement (similarity index [SI]) on ascale of no match to 0.965.The Sherlock microbial identificationsystem is solely based on computer comparison of the unknownorganism's fatty acid methyl ester profile with the profiles of apredetermined library of known isolates with pattern recognitionsoftware (Tang et al 1998).The fatty acid analysis by the MIDIsystem can be a useful supplement and reference method, butcannot be recommended at this time for the routine identificationof halophilic bacteria until a library of halophilic bacteriumisolates is established in the MIDI database.Presently, theMIDI system does not contain a library for halophilic bacteria.The database RTBSA6 is the closet database for halophilicbacteria (Chookiewattana , 2003). The data and isolate obtainedin this study can be used to develop the database in the MIDIsystem library for future identification of halophilic bacteria. Thehalophilic library can be constructed with ease as halophiles arestudied extensively throughout the world . Recently marine redpigmented Bacteria isolated from nellore costal region of andhraPradesh were studied for their fatty acid profile by 2.13analysis(pabba et al .,2011). Similarly the fatty acid methylesther profiles of 6 isolates belonging to bacteria domainrepresenting different genera and species were analyzed byGuvenk et al in 2010.V. CONCLUSIONTherefore, through the above study we can conclude thatFAME analysis and 16s rDNA sequencing are rapid and accuratemethods for bacterial identification .FAME analysis has anadded advantage of being cost effective as compared tosequencing. Thus this technology should be implemented bymicrobiologistsinto routine practice. However, the MIDIFAME libraries should be upgraded with chromatogram profilesof more micro-organisms. As halophiles from extremeenvironments like solar salterns of Goa, are potential candidatesfor enzymatic and other industrial applications, a library ofhalophilic microbes should be established.The halophilicmicrobial library can assist faster and accurate identification ofnovel halophiles.DescriptionCorynebacterium diphtheriaeCD450 16S ribosomal RNApartial sequenceCorynebacterium diphtheriaeCD449 16S ribosomal RNApartial sequenceCorynebacterium diphtheriaeCD448 16S ribosomal RNApartial sequenceMaxscoreTotalscoreMaxidentstraingene, 20620673%straingene, 20620673%straingene, 20620673%20673%Corynebacterium diphtheriae strainCD443 16S ribosomal RNA gene, 206partial sequenceCorynebacterium diphtheriae VA01,19597573%complete genomeCorynebacterium diphtheriae HC04,CP003215.119597573%complete genomeCorynebacterium diphtheriae HC03,CP003214.119597573%complete genomeTable 1: Blast Similarity Search Results fo H1 :Similarity to Corynebacterium diphtheriae strain CD450 by 16S ribosomalrRNAgene, partial sequence.CP003217.1www.ijsrp.org
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-315341.8001403.2600.7430.7502.700FID1 A, brarySim IndexEntry NameRTSBA6 6.000.596Corynebacterium 3.2123.2263.2761.8731.8981.9902.0152.071 2.0931.166601.5351.5681.07380m inFigure 1: Chromatogram of bacterial sample H1 showing the fatty acid peaks through Agilent GC bacillus pantothenticus strain OL0316S ribosomal RNA gene, partial sequenceVirgibacillus pantothenticus gene for 16SrRNA, partial sequence, isolate: T8-4TVirgibacillus pantothenticus gene for 16SrRNA, partial sequenceVirgibacillus pantothenticus gene for 16SrRNA, partial sequence, strain:PS11Virgibacillus pantothenticus gene for 16SrRNA, partial sequence, strain: NBRC102447Virgibacillus pantothenticus gene for 16SrRNA, partial sequenceVirgibacillus pantothenticus gene for 16SrRNA, 88%1447144788%Table2: Blast Similarity Search Results for H2:Similarity to Virgibacillus pantothenticus 16S ribosomal RNA gene, partialsequencewww.ijsrp.org
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-315350.7502.312FID1 A, LibrarySim IndexEntry NameRTSBA6 6.000.698Virgibacillus 1.4891.5691.188602.020 41004m inFigure 2: Chromatogram of bacterial sample H2 showing the fatty acid peaks through Agilent GC tBacillus atrophaeus strain BF 3fF 16S4732196100%ribosomal RNA gene, partial sequenceBacillus atrophaeus strain ZaK 16SJN824998.14731604100%ribosomal RNA gene, partial sequenceBacillus atrophaeus gene for 16S rRNA,AB681057.14732196100%partial sequence, strain: NBRC 16183Bacillus atrophaeus gene for 16S rRNA,AB680855.14732202100%partial sequence, strain: NBRC 15407Bacillus atrophaeus gene for 16S rRNA,partial sequence, strain: NBRC 13721AB680488.1 dbj AB680560.1 Bacillus atrophaeus 4732196100%gene for 16S rRNA, partial sequence,strain: NBRC 14117Bacillus atrophaeus strain LLS-M3-4 16SHM744708.14732191100%ribosomal RNA gene, partial sequenceBacillus atrophaeus strain KM40 16SJF411316.14732196100%ribosomal RNA gene, partial sequenceTable 3: Blast Similarity Search Results for H3:Similarity to Bacillus atrophaeus strain BF 3fF 16S ribosomal RNA gene,partial sequenceJQ693811.1www.ijsrp.org
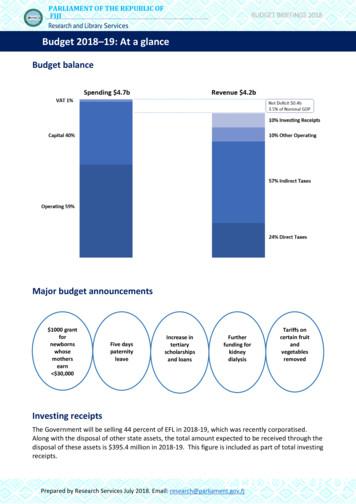
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-315362.8720.7322.259FID1 A, 2503.5m in4Figure 3: Chromatogram of bacterial sample H3 showing the fatty acid peaks through Agilent GC 6850.LibrarySim IndexEntry NameRTSBA6 6.000.776Bacillus Idiomarina zobellii strain G5B 16Sribosomal RNA gene, partial sequenceIdiomarina sp. TPS4-2 gene for 16Sribosomal RNA, partial sequence, strain:TPS4-2Idiomarina seosinensis strain PR58-8 16Sribosomal RNA gene, partial sequenceIdiomarina sp. TBZ1 16S ribosomal RNAgene, partial sequenceIdiomarina sp. JL974 16S ribosomal RNAgene, partial sequenceBacterium ABKPF4 16S ribosomal RNAgene, partial sequenceIdiomarina loihiensis strain GSP37 16Sribosomal RNA gene, partial 883511100%Table 4: Blast Similarity Search Results for H4:Similarity to Idiomarina zobellii strain G5B 16S ribosomal RNA gene, partialsequencewww.ijsrp.org
International Journal of Scientific and Research Publications, Volume 2, Issue 8, August 2012ISSN 2250-315370.7322.233FID1 A, (E12508.678\A0110007.
The technical resources necessary for sequence identification are significant. Despite the availability of . identifying the microorganism using online databases. The analysis data from the Sequencing . when it was applied to halophilic bacteria because their
inducted into both the Texas Black Sports Hall of Fame and the Texas High School Hall of Fame in 2004. He was inducted into the University of Colorado Hall of Fame in 2019, the Prairie View Interscholastic League Coaches Association Hall of Honor in 2014 and the California Sports Hall of Fame in 2018. Branch passed away in 2019 at the age of 71.
The Sanderson Athletics Hall of Fame was established by the Sanderson Athletic Club on November 8, 2010. The club’s Hall of Fame Committee is responsible for administering the Hall of Fame program. The purpose of the Sanderson Athletics Hall of Fame is to honor people who have distinguished themselves through extraordinary
Landry, and “Babe” Didrikson Zaharias, the Texas Sports Hall of Fame celebrates the athletes who have made a lasting impact on the the state. The 35,000-square-foot building also includes the Southwest Conference Hall of Fame, the Nolan Ryan Museum, the Texas High School Football Hall of Fame, and the Texas Tennis Hall of Fame. 9am - 5pm .
the NAIA Coaches Hall of Fame in 1973, and posthumously to the Kansas Sports Hall of Fame in 1977. He was a WWII veteran and was a charter member of the Braves Athletic Hall of Fame. The Peters Scholarship is a 1,000 award. Aaron Schoemann (Football) Schoemann is an Honorable Mention All-KCAC performer at defensive line for the Braves.
The Hall of Fame remained dormant until several prominent members of the Waco community created a plan in 1990 to have the Hall of Fame moved to Waco. Their plan was realized on April 16, 1993, when Waco had its grand opening for the Texas. Sports Hall of Fame. The museum also houses the Texas Tennis Museum and Hall of Fame. and Texas High .
Head Football Coach – 12 Years Career Record: 86-28-3 3 State Championships Inducted into the Bucknell University Hall of Fame Inducted into the Pennsylvania Coaches Hall of Fame Inducted into the Washington D.C Hall of Fame
The profile matrixfor a given motif contains frequency counts for each letter at each position of the isolated conserved region. 8 Sequence logo and consensus sequence We can extract the so-called consensus sequence, i.e. the string of most frequent letters: A graphical representation of the consensus sequence is called a sequence logo:
AngularJS Tutorial (SQLite) In this tutorial we turn to look at server-based web applications where the client-side code is AngularJS. Although a lot can be done with entirely browser-based (single-page) web applications, it is better to develop a server-based web application if any of the following are true: In-company (intranet) client machines may have restricted access to the Internet .