Pre-Lab E 5: Restriction Enzyme Digest And Plasmid Mapping (10 Pts)
Pre-Lab E 5: Restriction Enzyme Digest and Plasmid mapping (10 pts)Name: ; Lab Section: ; Grade:1. Describe the function of DNA ladder (1 pts).2.Describe the function of loading dye (1 pt).3. DNA fragments are charged, they will be drawn toward the electrode(1 pt).4. Which one is not the essential component of a restriction digest reaction? (1 pt)a) DNA plasmidb) Appropriate buffer (10X)c) Loading dyed) Restriction Enzyme5. What is the correct amount of agarose to make 55 ml of 0.75 % agarose gel? (showcalculation and unit, 2 pt.)6. The plasmid size of pGLO is 5371 bp and the restriction sites for for PstI are at 2106 and3181. Calculate to predict the sizes of fragments when pGLO is digested with PstI. (1pt)7. Short Answer: How is DNA visualized on agarose gel? (include reagents and equipments inyour answer) (2 pts)8. True or False: (1 pt, correct the errors to obtain full credits). In general, restriction sites are palindromic, meaning the sequence of bases readsthe same forwards as it does backwards on the same DNA strand.
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid MappingExperiment 5: Restriction Enzyme Digest and Plasmid MappingObjectives: By the end of this lab, you will be able to: Understand the use of restriction enzymes as biotechnology tools Become familiar with the principles and techniques of agarose gel electrophoresis Estimate DNA fragments sizes from agarose gel data Use a restriction map to predict how many fragments will be produced in a given restrictiondigest.Note: The introduction was adapted from Restriction Digest and Analysis of Lambda DNA Kit, Bio-RadLaboratories, Inc. Duplication of any part of the document is permitted for classroom use only.IntroductionThis excercise introduces you to some important principles of genetic engineering. Specifically, thefunctions of restriction enzymes and their use as molecular biology tools will be stressed. Usingagarose gel electrophoresis, you will examine the digestion patterns and determine the sizes ofunknown DNA fragments. Restriction enzymes were a catalyst for the molecular biology revolution,and now hundreds of such enzymes are commercially available. In this investigation, the restrictionenzymes EcoRV, and Pst I will be used to digest a plasmid, a small circular piece of DNA. Gelelectrophoresis will be employed to separate the resulting DNA fragments, and ethidium bromide willbe used to stain the DNA fragments for visualization.Restriction EnzymesThe ability to cut and paste, or cleave and ligate, a functional piece of DNA predictably and preciselyis what enables biotechnologists to recombine DNA molecules. This is termed recombinant DNAtechnology. The first step in DNA splicing is to locate a specific gene of interest on a chromosome. Arestriction enzyme is then used to cut out the targeted gene from the rest of the chromosome. Thissame enzyme is also used to cut the DNA of the recipient into which the fragment will be inserted.Restriction enzymes are proteins that cut DNA at specific sites. Restriction enzymes, also known asrestriction endonucleases, recognize specific sequences of DNA base pairs and cut, or chemicallyseparate, DNA at that specific arrangement of base pairs. They were first identified in and isolatedfrom bacteria that use them as a natural defense mechanism to cut up the invading DNA ofbacteriophages — viruses that infect bacteria. Any foreign DNA encountering a restriction enzymewill be digested, or cut into many fragments, and rendered ineffective. These enzymes in bacteria makeup the first biological immune system. Each restriction enzyme is named after the bacterium fromwhich it is isolated. For example:EcoRI The first restriction enzyme isolated from Escherichia coli bacteriaEcoRV The fifth restriction enzyme isolated from Escherichia coli bacteriaPstI The first restriction enzyme isolated from Providencia stuartii bacteriaEach restriction enzyme recognizes a specific nucleotide sequence in the DNA, called a restriction site,and cuts the DNA molecule at only that specific sequence. Many restriction enzymes leave a shortlength of unpaired bases, called a “sticky” end or “cohesive” end, at the DNA site where they cut,whereas other restriction enzymes make a cut across both strands creating double-stranded DNA1
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid Mappingfragments with “blunt” ends. In general, restriction sites are palindromic, meaning the sequence ofbases reads the same forwards as it does backwards on the opposite DNA strand. For example, here isa list of enzymes and the sites where they cut:EcoRI 5’G A-A-T-T-C 3’3’ C-T-T-A-A G 5’EcoRV 5’G-A-T A-T-C 3’3’ C-T-A T-A-G 5’PstI 5’C-T-G-C-A G 3’3’G A-C-G-T-C5’Setting up a simple restriction digest requires four mandatory ingredients: DNA: DNA that is free from contaminants such as phenol or ethanol. Excessive salt will alsointerfere with digestion by many enzymes. An appropriate buffer: different enzymes cut optimally in different buffer systems, due todiffering preferences for ionic strength and the type of cations. The restriction enzyme Deionized water: Specific amount of water is required to make up the final volume for a digestreaction.In this investigation, students observe the effects of two restriction enzymes on pGLO plasmid DNA.pGLO plasmid DNA is 5,371 base pairs, each restriction enzyme will cut the DNA one or severaltimes and generate restriction fragments of different sizes. In this activity, three separate samples ofplasmid DNA will be cut using two different restriction enzymes and the combination of them. Eachsample produces DNA fragments whose sizes can be estimated on an agarose gel electrophoresis.Electrophoretic Analysis of Restriction FragmentsIf a specific restriction site occurs in more than one location on a DNA molecule, a restriction enzymewill make a cut at each of those sites, resulting in multiple fragments of DNA. Therefore, if a givenpiece of linear DNA is cut with a restriction enzyme whose specific recognition sequence is found atfive different locations on the DNA molecule, the result will be six fragments of different lengths. Thelength of each fragment will depend upon the location of restriction sites on the DNA molecule.A DNA fragment that has been cut with restriction enzymes can be separated using a process known asagarose gel electrophoresis. The term electrophoresis means to carry with electricity. Agarose gelelectrophoresis separates DNA fragments mainly by size. DNA fragments are loaded into an agarosegel slab, which is placed into a chamber filled with a conductive buffer solution. A direct current ispassed between wire electrodes at each end of the chamber. Since DNA fragments are negativelycharged, they will be drawn toward the positive pole (anode) when placed in an electric field. Thematrix of the agarose gel acts as a molecular sieve through which smaller DNA fragments can movemore easily than larger ones. Therefore, the rate at which a DNA fragment migrates through the gel isinversely proportional to the log of the molecular weight (size or length). Over a period of time,smaller DNA fragments will travel farther than larger ones. Fragments of the same size stay togetherand migrate in single bands of DNA. A 500 bp ladder containing 16 bands in 500bp increments (thesmallest one is 500 bp) will be used to estimate the sizes of DNA fragments on a 0.8% - 1.0% agarosegel.Gels of 0.8-1.0% (w/v, 1 gram of agarose in 100 ml running buffer) agarose will separate fragmentsizes ranging from around 500 base pairs (bp) to around 10,000 bp (or 10 kilobases, kb). Largefragments (e.g., 10 kb) will be better separated in lower percentage agarose gel, such as a 0.5% gel.2
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid MappingOn the other hand, separation of very small (less than 1 kb) can be better achieved in higher percentageagarose gel, such as a 1.5-2% gel.Making DNA VisibleDNA is colorless so DNA fragments in the gel cannot be seen during electrophoresis. The loading dye(or loading buffer) does not stain the DNA itself but makes it easier to load the gels and monitor theprogress of the DNA electrophoresis. Loading buffer usually contains something dense (e.g. glycerol)to allow the sample to "fall" into the sample wells. It also contains one or two tracking dyes, whichmigrate in the gel and allow visual monitoring of how far the electrophoresis has proceeded. The dyefronts migrate toward the positive end of the gel, just like the DNA fragments. The “faster” dye,Bromophenol blue, comigrates with DNA fragments of approximately 300 bp, while the “slower” dye,Xylene cyanol, comigrates with DNA fragments approximately 9 kb in size. Visualization of the DNAsample requires the use of a transilluminator equipped with a UV lamp. A mid-wavelength (260-369nm) UV lamp emits light in the optimum range for viewing ethidium bromide-stained gels. You willview the gels while wearing safety glasses. This fluorescent dye intercalates between bases of DNAand RNA It is often incorporated into the gel so that staining occurs during electrophoresis, but the gelcan also be stained after electrophoresis by soaking in a dilute solution of ethidium bromide (EB). Youmust wear disposable gloves during the lab because EB is a mutagen and a suspected carcinogen. Youcan compare the DNA restriction patterns of the different samples of DNA when the bands are visibleusing UV lamp.In addition to electrophoresis, most of the procedures in this exercise employ microchemicaltechniques; i.e., using very small amounts of reagents such as DNA and enzymes. In researchlaboratory, many reactions of restriction digest require the addition of only one microliter (1 µl, oneone-millionth of a liter) of some components, so it is essential that you become familiar withmeasuring small volumes accurately and reproducibly.ProceduresMaterials and Equipments1) each work station ( for 2 students)a.b.c.d.e.f.g.h.i.4x loading dye (next to water bath)A yellow box of pipet tipsmicropipetter: p20a microcentrifuge tube rackmarkerThree microcentrifuge tubesA tube containing 30 µl of pGLO plasmid (0.03 µg/µl)One white container for used micropipette tipsdiluted EcoRV and PstI restriction enzymes in ice buckets at sink area2) every group (4 students will share one gel)a.b.c.d.e.f.g.Agarose (next to the balance)125 ml flask for melting agarose50 ml cylinder (near the sink)400 ml of 0.5XTBE buffer (near the sink)Gel apparatusPower supply10-teeth comb3
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid MappingNote: the gel apparatus comes as a set, i.e. the lid matches the tank. Clean the gel apparatus and slide thelid back onto the tank to prevent mismatches!1. Label three microcentrifuge tubes as follows:P (PstI digest)E (EcoRV digest)D (double digest of PstI and EcoRV, two enzymes pre-mixed)a) Transfer 10µl of pGLO plasmid to each tube. (check the volume setting on micropipette)b) Restriction enzymes have been pre-mixed with buffer by instructor and placed in ice buckets near frontsink. Using a red micropipette and a fresh tip for each tube, add 5 µl of diluted restriction enzymesaccording to the list above.c) Flick the tubes to mix well. Spin the tubes for 5 seconds in the microcentrifuge. The tubes should beplaced in a balanced configuration. Do not open the lid if the motor is still running.d) Incubate the tubes at 37ºC water bath for 50 minutes.* 1 unit of restriction enzyme activity is defined as the amount of enzyme required to produce a completedigest on 1µg of substrate DNA in 60 minutes at the appropriate assay temperature in a 50µl reactionvolume.2.[one gel per 4 students] Assemble the gel apparatus using a ten slot comb.a) Place gel casting tray on the top of gel support deck, the open ends of the tray should be placed nextto the sides of the chamber. Insert the comb into the end slot.b) Wear disposable gloves to prepare 50 ml of the melted 0.8% ( w/v, 0.8 gram/ 100 ml buffer) agarosesolution . The solution contains g of agarose and 50 ml of 0.5X TBE buffer (4.5mM Tris,4.5mM boric acid, 0.1 mM EDTA, pH 8.3). Boil the buffer and dissolve agarose completely usingmicrowave with 30 seconds intervals, the suspension needs to be completely clear.c) Ask TA to add 1µl ethidium bromide to your melted gel solution. Pour the gel solution to the castingtray. Rinse small flask with water.d) Demonstrations:e) After the solidification has occurred, lift the casting tray and rotate it 90 degree. Rinse the combwith water. Under most conditions, DNA has a negative charge caused by the phosphate groups onthe outer surface of the molecule. Negatively charged DNA molecules will migrate toward thepositive electrode during electrophoresis. You should place the end of gel with wells near(cathode [black] or anode [red]).f) Slowly fill the electrophoresis chamber with the 0.5X TBE buffer until the gel is completelysubmerged. The buffer should be about 3-4 mm above the gel surface.3. Retrieve your thee digested samples from the 37ºC water bath. Obtain one tube of 4X TBE loading dye(0.04% bromophenol blue, 20% glycerol, 0.04% xylene cyanol) for two students .4
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid Mapping4. (Your bench) Using a fresh tip for each digested sample, add 5 µl of 4X TBE loading dye. Flick the tubes tomix well and spin for 5 second in the microfuge.5. Set micropipette (p20) at 20 µl, load your three samples with fresh tips in the appropriate lanes following geldiagram Fig.1 as a guide. Do not leave space between each sample.6. Ask TA to load 5 µl of the 500bp DNA ladder (Bio-rad) and uncut pGLO plasmid in the middle lanes. TheDNA ladder contains 16 bands in 500bp increments; the smallest one is 500 bp.7. When the samples have been loaded, set the power supply to 130V. Run the bromophenol blue dye to within2 cm of the positive electrode end of the gel (35 minutes approximately). Predict the sizes of fragments inbase pairs based on the information in Fig 3.8. Visualize the DNA bands on a UV light box. Wear proper eye protection. You may trace the band pattern ona piece of plastic wrap overlaying the gel or take a picture using a digital camera.Note: The migration of DNA molecules in agarose gels is roughly proportional to the inverse of the log oftheir molecular weight (corresponding to the size of the fragments). The size of the DNA fragments can bedetermined by comparing the migration distances (mm) to the standard curve (log bp vs. migration distance)of the marker if the electrophoresis is run longer to achieve better separation. In this exercise, the length ofDNA fragments will be estimated based on the sizes of marker bands and the information provided by yourinstructor.Cleaning Up:1.2.3.4.Return the 4X loading dye tube to the tube rack next to water bath.Dispose the tips, sample tubes into big biohazard trashcan.Transfer the gel running buffer into the recycling bucket.Clean up the tank, the comb, the gel tray with tap water. Do not dry inside of gel box. Place the trayinside the tank and make sure the lid matches the tank.5. Refill yellow tip box.6. Wipe your bench with 70% ethanol (clear top squirt bottle).5
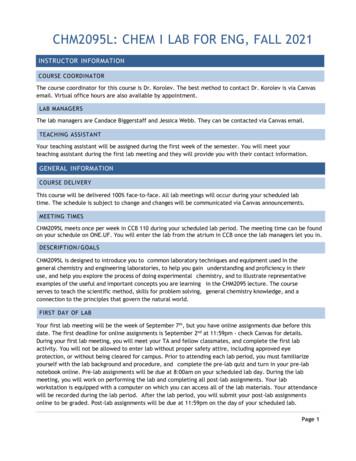
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid blePstI2000bp1500bp1000bp500bpFig. 1: gel diagramFig. 2: 500 bp DNA ladder5371 bpFig. 3: The map of pGLO : The restriction site for EcoRV is at 386 and the sites for PstI are at 2106and 3181)6
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid MappingPost Lab Report (20 pts total): Include questions and a title page in your report (1 pt).1. The restriction digest map of pUC19 is shown in Figure 4. Suppose you performed restrictiondigestions of pUC19 by the various enzymes listed in Table 1 (each row represents a separatedigestion),1) predict the sizes of fragments in base pairs and write your answers in Table 1(1pt);2) Label the positions of the wells (1pt), label the lanes with the enzymes used (2pt). Include a lanewith 500 bp DNA ladder (1pt, Fig 2). Draw the corresponding DNA fragments on the geldiagram, and label the size of each DNA fragment (2pts). Label the cathode and anode (1pt).0EcoR I 396Ava II 2059pUC19Ava II 18372686 bpBsey I 1110Figure 4. Restriction map of pUC 19, a 2,686 base pair plasmid. The number after each restrictionenzyme name indicates at which base pair the DNA is cut by that enzyme.Table 1.EnzymesFragments produced(bp)EcoRIBsey IAva IIEcoR1 Bsey IEcoR1 Ava IIBsey I Ava II7
Biol 2281, Spring 2016E 5: Restriction Enzyme Digest and Plasmid Mapping2. Print the photograph of your gel from eLearning. Label the wells, the lanes with the enzymesused (2pt). A 500 bp ladder containing 16 bands in 500bp increments (the smallest one is 500 bp)was used to estimate the sizes of DNA fragments on your gel. Note the sizes of the four smallestDNA fragments in the molecular ruler/ladder (1pt). Estimate and label the size of each fragmentin each restriction digest sample (1pt). Calculate the sizes of fragments in base pairs based onthe information provided in Fig 3 and label the size for each band (one set) (2pts). Label thecathode and the anode (1 pt).3. Visit the New England Biolabs website (www.neb.com) and click on “NEB CUTTER” at thebottom of the screen. Open the file of pGLO sequence found at eLearning. Copy and paste thesequence into NEBcutter. Make sure the sequence doesn’t have linebreaks in it. Type “pGLOyour name-BIOL2281” in “Name of sequence”. Select “the sequence is circular”. Select NEBenzymes and click submit. You will get the restriction map for pGLO.Then click on Custom Digest under Main Options. Choose EcoRV, PstI. Select “Digest”. Itwill display the restriction map with just these enzymes. Click “view gel” under “Main options”.Print the page with a virtual gel and the list of fragments. Your name should be displayed on thetitle of the print-out (2pts).On the page of “Custom Digest”, five of the open reading frames are identified on the plasmidmap. Click on the curved bars of c. Click “Blast this sequence at NCBI” under the proteinsequence and find out the name of the protein superfamily (1pt). Repeat the search steps forcurved bar of b and find out the name of the protein superfamily (1pt).8
BIOL2281 E5Restriction Enzyme DigestExperiment 5: RestrictionEnzyme Digest and PlasmidMapping Restriction Enzyme DigestPlasmid MappingGel electrophoresis A strategy for obtaining fragments of DNA Restriction enzymes cleave segments of DNA fromthe genome of various types of cells or fragmentDNA obtained from other sources Restriction fragments of DNA from different sourcescan be used to synthesize a recombinant DNA12Types of nucleases Nucleases: catalyze the hydrolysis of phosphodiesters innucleic acids Exonucleases: from one end of a polynucleotide chainEndonucleases: at various sites within a polynucleotidechain Restriction Endonucleases (Enzymes):synthesized in some bacteria to protect against viralattack by destroying foreign DNAHow do they protect their own DNA? The host cell modify the basesof potential restriction sequences by methylation of adenine orcytosineDNA cloning: The new plasmid can be introduced into bacterialcells that can produce many copies of the inserted erting.htmlRestriction recognition sequencesNuclease cleavage sites Restriction Enzymes (RE) cleaves the DNA within or near to thespecific recognition sequencesthe nucleotide sequences recognized by more than 200 RE can beclassified as:a. Tetra- or hexapalindromic sequences:they are the same sequences when read in the5' --- 3' didirectionti off bbothth strands.t dDpn I 5’GATC3’EcoR I 5’GAATTC3’3’CTAG5’3’CTTAAG5’b. Pentanucleotide sequences.c. Sequences of longer extention with internal (N) sequences.Xmn I (GAANNNNTTC)d. Nonpalindromic sequences.5Dr. Wenju Lin, BIOL2281 Spring 2016(Nawin Mishra,2002)61
BIOL2281 E5Sticky and Blunt ends78Figure 8.3Essential components of arestriction digest reactionMicroscale DNA plasmid Appropriate buffer (10X) dH2O Enzyme (unit/µl) : the volume of enzyme should not beincluded in final reaction volumeThe volumes and tools are adapted to microscalewhen we work with DNA 1 ml 1000 µl (1000 microlitres)1 mg 1000 µg (1000 micrograms) Microcentrifuge tubes0.25 ml to 2.0 mlMade of PolypropyleneWith Flat tops for writing onWith Graduations, and writing area on theside of the tube.One unit of restriction enzyme activity is defined as the amount of enzymerequired to produce a complete digest on 1µg of substrate DNA in 60minutes at the appropriate assay temperature in a 50µl reactionvolume. Most restriction enzyme reactions are incubated at 37 Cfor one hour.9Microcentrifuge Electrophoresis Use centrifugal force to separatelighter (rich medium) and heaviersubstances (cells).Spin tubes to ensure efficient usage ofvaluable drops of reagent. a separation technique in which an electricalfield causes charged molecules to movethrough a matrix (usually a gel).routinelyy used to separatepDNA,, pprotein andother polymeric molecules.Separation can be based on Use pause button for momentary spin The load in a laboratory centrifugemust be carefully balanced.Dr. Wenju Lin, BIOL2281 Spring 201610 11Sizes (DNA fragments separated by size).Net chargesshapes122
BIOL2281 E5The EquipmentsElectrophoresis Equipments & Setup The tray The comb: The actual mold which provides a shape for the gel .It is placed into slots in the tray, with the "teeth" down, when theagarose is still hot. The agarose polymerizes with small "wells“ intowhich samples are added.The tank Holds the running buffer, the same buffer used to make the gelis also used as the running buffer. One power connection lead is positive (red) and one negative,there will be a strong electrical current flowing through the tankwhen the electrodes are immersed. The gel will be completelysubmerged as it is run.1. The flat-bed tank2. The gel tray3. The comb13Visualizing DNA byEthidium BromideAgarose gel electrophoresis Agarose (nontoxic polysaccharides) gel 14Mixed with gel running bufferMeltedPoured into a casting trayDNA ffragmentst are charged,hd ththey willill beb drawndtowardtd thethelectrode (anode,red). DNA fragments are separated by. DNA fragments stained by ethidium bromide (EB) are visible usingUV lamp. EB Intercalates into the DNA molecules and is acarcinogen. The lengths of the DNA fragments can be determined by comparingthe migration distances (mm) to the standard curve (log bp vs.migration distance) of the DNA ladder.15 1 g/ml of EB included inthe agarose gel or TBEbuffer16Effect of DNA Conformation onMobility500bp DNA ladder Contains 16 bands in 500bpincrements; the smallestone is 500 bp Used to estimate DNAfragment sizes on a 0.8% 1.0% agarose gel.wells The size of uncut plasmid DNA can not be accuratelydetermined. Uncut plasmid DNA has several distinctconformations. CircularCil fform vs lilinear DNASupercoiled vs nicked circles; 1000 bp The mobility of linear DNAfragments is inverselyproportional to the log10 oftheir molecular weight.500 bp17Dr. Wenju Lin, BIOL2281 Spring 2016183
BIOL2281 E5Agarose gel electrophoresis (con.) Use of Restriction mappingThe total size of the plasmid can beobtained by adding up the size ofeach DNA band in one lane A description (roadmap) of restrictionendonuclease cut sites within a region of DNA To characterize an unknown DNA priorsubcloning or further manipulations, ex:recombinant plasmid mapping DNA fingerprinting, ex: restriction digestions onPCR products, RFLPPartial digests Intensity of DNA bands correlate tothe amount or the molecular numbersof DNA19Single digests and doubledigestsPlasmid Mapping Cutting a plasmidconverts a circularmolecule into a linear one. 20Cutting a plasmid withtwo enzymes results intwo linear fragments. Single digests are used to determine whichrestriction sites are in the unknown DNA or todetermine the size of a plasmid. Double digests are used to order and orient thefragments correctly.Enzyme 1Enzyme 2How about cutting a linear PCR fragment?2122Draw what you would see on a gelwith the predicted fragmentsPredict fragment size based onplasmid mapSingle digest 0EcoR I396 EcoRI- 2686bpBsey IAva IINegativeelectrodeAva II2059pUC 19Ava II18372,686 bpBsey I1110Double digest EcoRI Bsey I1110-396 714,2686-714 1972 EcoRI Ava II Bsey I Ava IIEcoR I Bsey I7142686DNA Markerwell23Dr. Wenju Lin, BIOL2281 Spring 20161972EcoR I500 bp244
BIOL2281 E5The map of pGLO38653713181 2106The restriction site for EcoRV is at 386 and the sites for PstI are at 210625and 3181)The concentration of an agarose gel allowsfor the separation of different sizes of DNAfragments.What factors influence the rate ofmigration in DNA agarose gel? 26Molecular size of the DNAThe conformation of DNAThe agarose concentrationThe bufferThe applied voltage 0.5% gel : providing better separation for fragmentslarger than 10 kb 1% ggel: providing better separation for fragmentsbetween 500bp-10kb 2% gel: providing better separation for fragmentssmaller than 1 kb 0.8% gel will be used in the lab27Loading DyeVoltage Used to monitor the movement of DNAfragments, it does not stain DNAThe higher the voltage, the faster the rate ofmigration. However, 28Accompanying heat may melt low-percentage gel.(Don’tt use more than 130 volts)(Donvolts).Imperfections in the gel distort the bands andproduce ambiguous results (slants and smiles).TBE loading dye contains Glycerol Bromophenol blue: migrates as a 300bp fragment. Xylene cyanol: migrates as a 9000bp (9Kb) fragment. 29Dr. Wenju Lin, BIOL2281 Spring 201610X stock routinely added to the samples beforeloadingIn the lab: 15µl of reaction mix 5µl of 4X loading dye305
BIOL2281 E5Restriction fragments cut with twoenzymes with complementary tailsIn the lab Complete restriction enzyme digests of pGLOSet up agarose electrophoresisComplete Question1 in report E5Photo of gels will be posted on eLearningAnalyze the plasmid further at home byvisiting the New England Biolabs website andNCBI website31Dr. Wenju Lin, BIOL2281 Spring 2016Sal IG* TCGA CC AGCT *GGGXHO IC* TCGA GG AGCT *CCCCAGCTCAGCTGAGCTGAGCTTCGACTCGAC GG TCGAGTCGAGCC326
Restriction Digest and Analysis of Lambda DNA KitThe introduction was adapted from , Bio-Rad . laboratory, many reactions of restriction digest require the addition of only one microliter (1 µl, one one-millionth of a liter) of some components, so it is essential that you become familiar with
Penny's SEP:SEP2006CD:Gelelectrophoresis:DNA Lab #206.doc 7/19/06 11:07 AM 2.1 LAB 2: RESTRICTION ENZYME DIGESTION (CUTTING) OF DNA In Lab 1, you worked with an essential research method widely used in genetic analysis and biotechnology. The DNA in your samples had already been cut by restriction enzymes. In this lab,
Each week you will have pre-lab assignments and post-lab assignments. The pre-lab assignments will be due at 8:00am the day of your scheduled lab period. All other lab-related assignments are due by 11:59 pm the day of your scheduled lab period. Pre-lab assignments cannot be completed late for any credit. For best performance, use only Firefox or
Biology Lab Notebook Table of Contents: 1. General Lab Template 2. Lab Report Grading Rubric 3. Sample Lab Report 4. Graphing Lab 5. Personal Experiment 6. Enzymes Lab 7. The Importance of Water 8. Cell Membranes - How Do Small Materials Enter Cells? 9. Osmosis - Elodea Lab 10. Respiration - Yeast Lab 11. Cell Division - Egg Lab 12.
Contents Chapter 1 Lab Algorithms, Errors, and Testing 1 Chapter 2 Lab Java Fundamentals 9 Chapter 3 Lab Selection Control Structures 21 Chapter 4 Lab Loops and Files 31 Chapter 5 Lab Methods 41 Chapter 6 Lab Classes and Objects 51 Chapter 7 Lab GUI Applications 61 Chapter 8 Lab Arrays 67 Chapter 9 Lab More Classes and Objects 75 Chapter 10 Lab Text Processing and Wrapper Classes 87
KEY CONCEPT Biotechnology relies on cutting DNA at specific places. VOCABULARY restriction enzyme restriction map gel electrophoresis MAIN IDEA: Scientists use several techniques to manipulate DNA. 1. List five ways in which scientists study and manipulate DNA. MAIN IDEA: Restriction enzymes cut DNA. 2. What is a restriction enzyme? 3.
Restriction Mapping 6. A circular DNA plasmid, pDA102, has a size of 4.35 kb. When the plasmid DNA digested with combinations of restriction enzymes and the resulting fragments are electrophoresed, the following data is obtained. Using these data, construct a restriction map of pl
LABORATORY3 DNA Restriction Analysis L ABORATORY 3 INTRODUCES THE ANALYSIS OF DNA using restriction enzymes and gel electrophoresis. Three samples of purified DNA from bacteriophage λ (48,502 bp in length) are incubated at 37ºC, each with one of three restriction endonucleases: EcoRI, BamHI, and HindIII. Each enzyme has five or more restric-
Despite the infiltration of over 20,000 words from English into the Japanese language, has the Japanese language disappeared? Has the grammatical structure or pronunciation of Japanese changed dramatically since World War II? Schwarz and Ezawa introduce the Japanese language to foreigners as they demonstrate Japanese culture to Westerners when they write “what is a mikoshi?” They further .