Gabriella Miller Kids First Pediatric Research Program
Gabriella Miller Kids FirstPediatric Research ProgramPublic WebinarOctober 6, 20202:00 pm EDT
Webinar Agenda2:00pm - Introduction; NIH Kids First Staff2:05pm - Progress on the Kids First Study on NovelCancer Susceptibility in Families; Dr. Sharon Plon, BaylorCollege of MedicineoQuestions for Dr. Plon2:35pm - Kids First Data Resource Center;Kids First DRC StaffoNew Portal FeaturesoKids First Variant Workbench Demo3:05pm - NIH Common Fund Data Ecosystem (CFDE)presentation; Dr. C. Titus Brown, UC Davis3:25pm - NIH Program Updates; NIH Kids First Staff3:40pm - Questions & Answers
Valerie CottonKids First Program ManagerEunice Kennedy Shriver National Institute ofChild Health and Human Development (NICHD)
NIH Kids First Working GroupKids First is an NIH Common Fund program coordinated by a trans-NIH WorkingGroup, which is chaired by four institutes:Eunice Kennedy Shriver National Institute ofChild Health and Human Development (NICHD)National Human Genome Research Institute(NHGRI)National Heart, Lung, and Blood Institute(NHLBI)National Cancer Institute(NCI)Other Working Group MSNIAIDNCATSORIPCDC
How did Kids First get started? Initiated in response to the 2014 Gabriella Miller Kids FirstResearch Act:––––Signed into law on April 3, 2014Ended taxpayer contribution to presidential nominating conventionsTransferred 126 million into the Pediatric Research Initiative FundAuthorized appropriation of 12.6 million per year for 10 years to theNIH Common Fund for pediatric research; first appropriation was forFY2015
VisionAlleviate suffering from childhood cancerand structural birth defects by fosteringcollaborative research to uncover theetiology of these diseases and supportingdata sharing within the pediatric researchcommunity.
Why study childhood cancer & structural birthdefects together? Both are leading causesof childhood mortality Birth defects areassociated withincreased risk of canceramong children suggesting sharedgenetic pathwaysFrom: Association Between Birth Defects andCancer Risk Among Children and Adolescentsin a Population-Based Assessment of 10Million Live BirthsLupo et al, JAMA Oncol. 5
Kids First Major InitiativesThrough 2021:1. Identify & sequence cohorts of children with childhood cancer and/orstructural birth defects.2. Build the Gabriella Miller Kids First Data Resource to empower discoveryYear 15 16 17 18 19 20 21 22 23 24Sequencing (U24)Phase 2
The Kids First Dataset is Growing!40 projects 40,000 genomes 16,000 cases 14 released datasets Disorders of Sex DevelopmentCongenital Diaphragmatic HerniaEwing SarcomaStructural Heart & Other DefectsSyndromic Cranial Dysinnervation DisordersCancer SusceptibilityAdolescent Idiopathic ScoliosisNeuroblastomasEnchondromatosesOrofacial Clefts in Caucasian, Latin American,Asian & African, Filipino populationsOsteosarcomaFamilial LeukemiaHemangiomas, Vascular Anomalies &OvergrowthCraniofacial MicrosomiaIntersection of childhood cancer & birth defectsMicrotiaEsophageal Atresia and TracheoesophagealFistulas Kidney and Urinary Tract DefectsNonsyndromic CraniosynostosisBladder ExstrophyHearing LossCornelia de Lange SyndromeIntracranial & Extracranial Germ Cell TumorsFetal Alcohol Spectrum DisordersMyeloid Malignancies overlap with DownsyndromeCongenital Heart Defects & Acute LymphoblasticLeukemia in Children with Down SyndromeStructural Brain DefectsStructural Defects of the Neural Tube (SpinaBifida: Myelomeningocele)CHARGE SyndromeLaterality Birth DefectsT-cell Acute Lymphoblastic LeukemiaPediatric RhabdomyosarcomaValvar Pulmonary Stenosis
The Kids First Data Resource for Collaborative DiscoveryData Resource PortalKnowledge BaseIntegrations(PedcBioPortal)Entry point. Query, search, discover, build &visualize synthetic cohortsIntegrations with existingcurated/published data visualizationsData ServicesModel clinical data inFHIR-based dataservices for semanticinteroperability andcoordinationCavaticaPull data from multiple sources intoone workspace.Use notebooks, bring-your-own oruse available workflows.Framework ServicesIndex and point to files in thecloud (for approved users)STRIDES
More researchers are accessing Kids First data! 1700registeredusers since2018 launchIndividual-level sequence data 150 Data Access Requestsapproved by the Kids First DataAccess Committee across 14 KidsFirst genomic datasets availableNIH Kids FirstData Access Committee
Researchers are using Kids First data toanswer new scientific questions 14 awards for R03 for analyses of Kids First data(PAR-16-348 ; PAR-18-733; PAR-19-069, PAR-19-375) 2 awards for NIDCR R03 (PAR-16-070) 2 awards for INCLUDE R03 (RFA-OD-20-006) 3 awards for R01s (PA-13-302, PAR-17-236) Spurred new collaborations with KOMP2 & INCLUDEKnockout MousePhenotypingProject (KOMP2)INvestigation of Co-occurringconditions across the Lifespanto Understand Downsyndrome (INCLUDE)
Researchers Are Making Discoveries19 Journal Publications To-DateAverage RCR: 1.36https://commonfund.nih.gov/publications?pid 40
Kids First X01: Identifying novel cancer susceptibility mutationsfrom unselected childhood cancer patient and parent triosSharon E. Plon, MD, PhD, FACMGBaylor College of MedicineOwen HirschiBaylor College of Medicine
1 X01 HL136994-01 - Identifying novel cancersusceptibility mutations from unselectedchildhood cancer patient and parent trios (BASIC3)Sharon E. Plon, MD, PhD, FACMGDepartments of Pediatrics/Hematology-Oncologyand Molecular and Human GeneticsHuman Genome Sequencing CenterBaylor College of Medicine
Disclosures – Sharon E. Plon, MD, PhD I have the following financial relationshipsto disclose:– I am a member of the Baylor GeneticsLaboratory Scientific Advisory Board
Three different approaches to understandinggenetic basis of childhood cancer Several studies of clinical genome-scale testing of diversecohorts of childhood cancer patients (BASIC3 and TexasKidsCanSeq study) Sequencing of large cohorts of specific pediatric cancerpatients Precision oncology treatment trials of matched tumor/normalsequencing for patients with relapsed or recurrent tumors(NCI/COG Pediatric MATCH Trial)
Baylor College of Medicine BASIC3 Key Team MembersWill ParsonsMuraliPediatric Oncology SueHilsenbeckAngshumoyRoyDoloresLopez-Terrada
3BASICBaylor College of Medicine Advancing Sequencing Into Childhood Cancer CarePATIENTS280 children newly diagnosedCNS and non-CNSsolid le exomesequencingRETURN OF ilyNo relapseMUTATIONREPORTSStudy objectives: To integrate information from CLIA-certified germlineand tumor exome sequencing into the care of newlydiagnosed solid and brain tumor patients at TexasChildren’s Cancer Center To perform parallel evaluation of the impact of tumorand germline exomes on families and physiciansWill ParsonsPediatric Oncology
Race/Ethnicity of BASIC3 Subjects are Representativeof Houston Population6% Other38% Latino11% Black45% AngloTexasCharacteristics of patients enrolled and not enrolled on study - updatedEnrolledDeclinedCharacteristic - no. (%)P Value(n 239)(n 103)Ethnicity0.54HispanicNon-HispanicNot reportedRace111 (46%)119 (50%)10 (4%)WhiteBlack or African AmericanAsianAmerican Indian or Alaska NativeMultipleNot reported141 (59%)25 (10%)7 (3%)10 (4%)14 (6%)42 (18%)41 (40%)52 (50%)10 (10%)0.1174 (72%)12 (12%)4 (4%)2 (2%)---Updated from Scollon et al., Genome Medicine 2014
BASIC3 DIVERSE PEDIATRIC TUMOR DIAGNOSESNON-CNSCNS81/94 (86%)40/56 (71%)Tumor available for WES
Diversity of clinical exome results returnedTUMORREPORTAllGERMLINEREPORTGeneCancer or OtherPatient nsenseRare WT1missenseSCN5AmutCYP2AmutCFTR F508Opt-In
Cancer susceptibility molecular diagnosis in 9.8%(27/278) pediatric cancer patientsAutosomal dominant 26(P/LP)19 different genesGenes associated w/specific childhoodcancer15Examples include DICER1,VHLx3, MSH2, WT1x2,TP53x3Genes not previouslyassociated w/specific childhoodcancer11Examples include BRCA1x2,BRCA2, PALB2, CHEK2x2,FLCN, SMARCA4Autosomal recessive 1(biallelic)TJP2No one gene was reported in more than 3 BASIC3 patients:3 each for VHL and TP53.
Rationale for WGS from Heterogeneous Dataset Several examples from BASIC3 exome data of identifying arare variant in a single individual that was then enlarged toidentify other rare cases outside the study resulting in newgermline or somatic drivers.– TJP2 deficiency and risk of hepatocellular carcinoma– Internal tandem duplication of BCOR as major somatic driver of clearcell sarcoma of the kidney
Goal of KidsFirst Project Utilize the rich existing dataset from BASIC3 to make furtherdiscoveries using whole genome sequencing of proband andparent trios. Existing data:–––––Germline exomes of probands onlyTumor exomes from probandsRNAseq for a subset of tumorsDNA from patient and parents isolated from blood samplesLymphoblastoid cell lines available from almost all patients andparents for subsequent functional or splicing studies as needed.
Approach to KidsFirst Analysis Perform sequencing of patient/parent BASIC3 trios withfollowing priority:– Complete trios with adequate DNA– Patients with unusual histories (multiple malignancies, birth defects)– Some solved cases to look for evidence of genomic instability For example, number of de novo variants in child of TP53 carrier Analysis has focused on both single nucleotide variants (SNV)and structural variants (SV)– Prioritized de novo variants in both situations– Also analyzed known cancer susceptibility genes for “missed” variantsfrom exome analysis– Performed spliceAI analysis of rare variants for cryptic splicing variants
De novo SNV calling in BASIC3 WGS trios (n 54complete trios)Prioritization:PlatypusOutcome:
De novo heterozygous missense variant in KMT5Cin pediatric ependymoma Mutation is rare, occurs in highly conserved functionaldomain, and has a high CADD (28.3) and REVEL (.662)scoreKMT5C
STRUCTURAL VARIANT CALLING INBASIC3/KIDSFIRST WGS DATAOwen HirschiGraduate StudentGenetics and Genomics ProgramBaylor College of Medicine
Identification of germline structuralvariation in childhood cancer is limitedSomatic structural variants play a critical role in cancersChr. 22Chr. 9Ex. Philadelphia chromosome translocation in leukemiaGermline structural variants are implicated in pediatric cancersAPCDiscovered throughRNA-seqEx. Inversion of a cancer-associated gene ina child with liver cancerHaines K, et al. 2019Since 2010 over 40 programs have been created to identify SVfrom short-read WGS - primarily built for identification in tumors Sensitivity ranges from 10-70% and false positive rate up to 89%Alkan C, et al. 2011Mahmoud M, et al. 2019
Developed new pipeline for more effectivegermline structural variant callingCaller A, B, C, D, & E:Lumpy, Manta, Delly,Breakdancer, & CNVnatorUsing multiple structural variant callers allowed for morespecificity and sensitivity in variant identificationThe pipeline is built for identification of de novo structuralvariants and is being tuned for inherited variantsHurley Li, PhD
Analysis of structural variant on Cavatica requires multiplefeatures of the platform
De novo structural variant calling in BASIC3 WGStrios (n 54 trios)Analysis:Outcome:
Germline de novo heterozygous 20kbp deletion ofNF2 in a meningioma patientFatherMotherProbandPotentially SomaticMosaic6/40 readsNF2 exon:5-9
Germline de novo heterozygous 8kbp duplicationof PTGR2 in an ependymoma R2 promoter and exon 1
PTGR2 duplication includes 5 predicted erENCODE
Whole genome sequencing revealed novel germlinestructural variantsWe have developed a pipeline with the aim to betteridentify germline de novo structural variation in pediatriccancerWe have identified a germline de novo heterozygousduplication in a pediatric ependymoma that we arebeginning to functionally assessWe have benefited from additional research studiesinvolving both tumor and germline sequencing of BASIC3
Long-read sequencing of BASIC3 through KidsFirstLong-read sequencing allowsfor greater detection of SVStructural Variants ObservedLong-readsShort-readsGoals of this sequencingstudy:We will compare the SV results of the long-read sequencing to those of theshort-read sequencing using our pipelineEnabling us to identify: Limitations and accuracy of short-read structural variant in trio studies Causative SVs only found via long-read sequencingMerker JD, et al. 2017
QUESTIONS?
Gabriella MillerKids First DataResource Center
Adam Resnick, PhDChildren’s Hospital of PhiladelphiaPrincipal Investigator,Gabriella Miller Kids FirstData Resource CenterVincent Ferretti, PhDSainte-Justine University HospitalPrincipal Investigator,Gabriella Miller Kids FirstData Resource Portal
KFDRC Portal Update
Methodology45
Double “funnel” model46
47
Released StudiesStudy NameFunding YearCategoryParticipantsTriosOther l Diaphragmatic HerniaFY15 & FY16 &FY17Structural Birth Defects224571460Congenital Heart Defects (PCGC)FY15 & FY16Structural Birth Defects2133711n/aOrofacial Clefts - EuropeanAncestryFY15Structural Birth Defects129538090Orofiacial Clefts - Asian & AfricanAncestryFY17Structural Birth Defects7252386Novel Cancer Susceptability(from BASIC3)FY16Cancer291664848
Released Studies (Cont’d)Study NameFunding YearCategoryParticipantsTriosOther FamiliesEwing SarcomaFY15Cancer1047290153Syndromic CranialDysinnervationFY15Structural Birth Defects801248n/aOrofacial Cleft - Latin AmericanAncestryFY16Structural Birth Defects8042629Adolescent Idiopathic ScoliosisFY16Structural Birth Defects262657Familial ural Birth Defects79252Disorders of Sex DevelopmentFY15Structural Birth Defects300913OsteosarcomaFY15Cancer84proband-only studyn/a49
Approaching 30,000 samples released and in the que 2000 3000 1000 3500 100 300 2500 3000 500 400 2500 1000 800 300 1600 300 100050
51
The Kids First Data Resource for Collaborative DiscoveryData ResourcePortalKnowledge BaseIntegrations(PedcBioPortal)Entry point. Query, search,discover, build & visualizesynthetic cohortsIntegrations with existingcurated/published datavisualizationsDataServicesCavaticaPull data from multiplesources into one workspace.Use notebooks, bring-yourown or use availableworkflows.Model clinical data inFHIR-based dataservices forsemanticinteroperability andcoordinationFramework ServicesSTRIDESIndex and point to files in thecloud (for approved users)
Ontologies Enable Computable Modelshpo.jax.org
Curation of Kids First data setsEngage withcommunity foradditions and/orrecommendations
55
Variant Workbench
Double “funnel” model57
Interoperability Across NIHResourcesFramework ServicesIndex and point to files in thecloud (for approved users)
Kids First DRC Team
Kids First DRC Team
The KF Germline Variant Workbench Project Prototype presented at the last webinar in May Productionizing software and infrastructure Security, data quality, robustness, bioinformatics, clustermanagement and software deployment tools Objective Extract, annotate and import variant data from harmonized gVCF filesto a queryable database Challenges Complex data, big data, confidentiality and security Production version to be released Phase I: X01 investigator’s teams, Oct-Nov 2020 Phase II: All Kids First Portal members, Jan 2021
Two main components An annotated variant database User private compute environment with Spark and ZeppelinHarmonizedgVCFsParticipantclinical dataPublicvariantdatasetsE.gTOPMedKF VariantDatabaseParquet fileson AmazonS3User’s ZeppelinNotebooksUser’s terClusterCluster
Variant DatabaseData is organized into tables like in a traditional relational database31 tables in current man genesclinvarbiospecimenparticipantsvariantstopmed bravodiagnosesPhenotypesconsequences.
Current Annotations From VEP and dbsnfp AA change, genes, transcripts,. 360 different annotations (e.g. predictive functionalscores, gtex) Gene ids and alias from the NCBI Clinvar, dbSnp Gene panels Cosmic, HPO, Omim, Orphanet Populational databases TOPMed, gnomAD 2.1 and 3.0, 1000genomes KF annotations Allele frequencies, parent’s genotypes,zygosity, QC metricsExample: Find all de-novo rare missense variants with high functional impact in low grade gliomapatients affected by any cardiovascular abnormalities
Data Currently Loaded for Initial Beta Phase11 studies11 thousand participants297 million unique variants53 billionoccurrences Each variant occurs on average in 178 participantsEach participant has on average 4.8 million variants52% (153M) of the KF variants are in TOPMed and/orgnomAD
The Zeppelin Data Analytic EnvironmentProvides programmatic access to the variant database from webbrowsers Accessible from the Portal Powered by individual Sparkclusters on AWS User notebook workspace forcreating and managingnotebooks Users can only access variantdatasets they have beenauthorized to
Zeppelin Notebooks Support for various programmaticlanguages (SQL, Python, R, Scala,Markdown) Can use different languages in the sameNotebook Built-in chart plotting capability Interactive forms Multi-user capability Pre-installed libraries Hail, Pandas, Numpy, Matplotlib,Plotly, Seaborn
Cohort builder & variant database integrationVirtual cohorts built using the Portal cohort builder can be saved as Participant setsUser dashboard
User’s saved sets as part of the variant databaseSaved sets can be joined with other variant tables for genomic analysesof virtual cohortsVariant workbench
Who is the variant workbench for? Initially: KF DRC for data quality control Bioinformaticians who know programming in SQL, Python, R, etc. Next Aim: Increase the user base by offering a series of interactive andcomprehensive notebooks directly accessible from the portal e.g. a gene-centric analysis notebookUsers will be able to input analysis parameters (Zeppelin forms)import their own data to their notebooks (e.g. gene panels, variant datasets)modify notebooks as they need and save them in their workspaceA good tradeoff between standard data portal static analyses and the complexity ofprogramming notebooks from scratch
Further workIndexing the variant database with ElasticsearchBuilding data querying interfaces within the portal (integrating notebook use cases) GA4GH-like Beacon service that returns yes/no answers on variantoccurrences GnomAD-like interface that returns aggregations e.g. allele frequencies Integration with the Cohort Builder enabling complex queries with bothclinical and genomic criteriaAdding more annotations supporting variant prioritizationKF VariantDatabaseParquet fileson AmazonS3ElasticSearch ClusterData indices
Special Thanks ToCHU Ste-Justine Jeremy Costanza, Lead software architect and developerDevelopers Adrian Paul, Evans Girard,Francis LavoieUX Lucas LemonnierCHOP DevOps Alex LubneuskiBioinformatics Yuankun Zhu, Yiran Guo, Miguel Brown, David Higgins
COMMONFUND DATAECOSYSTEMAN OVERVIEWOCTOBER 6TH, 2020C. TITUS BROWN, UC DA
Baylor College of Medicine. Owen Hirschi . Baylor College of Medicine. . and tumor exome sequencing into the care of newly . Diversity of clinical exome results returned. Cancer susceptibility molecular diagnosis in 9.8% (27/278) pediatric cancer patients. Autosomal dominant
Communication Skills Learning Tools for the Pediatric Clerkship 37 Pediatric History Taking Approach to the Pediatric Patient 38-39 Explanation of Pediatric H&Ps/Pediatric Database 40-43 Example H&Ps (older child and infant) 44-52 Pediatric Physical Examination Benchmarks for Pediatric Physical Examination 53 54-65
MILLER Syncrowave 350LX TIG Welder, 350-AMP @ 40% w/Miller Cooler MILLER Shopmaster TIG Welder, 300-AMP @ 50% w/Miller Cooler MILLER Dialarc HF TIG Welder, 250-AMP @ 40%, Miller Coolmatic 3 MILLER DX Portable TIG Welder (11) MILLER Dialarc 250 AC/DC Stick Welders, 225-AMP @ 30% (3) LINCOLN Idealarc 250 Stick
The physicians at Albany Med's Bernard & Millie Duker Children's Hospital are specially trained in more than 40 pediatric fields, including pediatric pulmonary disease, pediatric surgery, pediatric gastroenterology, pediatric anesthesia and pediatric neurology. Albany Med houses the region's only Pediatric Intensive Care Unit (PICU) and
(10) Late Model Miller and Lincoln Tig Welders; (2) Lincoln Precision Tig 375, (2) Miller Dynasty 350’s, (2) Miller Syncrowave 350LX’s, Miller Dynasty 300DX, Miller Syncrowave 250, (2) Miller Dynasty 200’s SEE PHOTO (8) Miller Spectrum 2050 Plasma Cutters SEE PHOTO Large Quantity of W
Pediatric hematology Organizing comprehensive cancer care for children Pediatric oncology Pediatric brain tumor Forming working groups with pediatric hematologists & pediatric oncologists Drafting National Cancer Control Plan - engaging pediatric hematology & oncology & WHO/IARC/IAEA/St. Jude experts Defining national needs for cancer workforce
Ophthalmology MidValley 77 - 79 Ophthalmology Pediatrics 80 Optometry OVMC 81 Optometry MidValley 82 . Speciality Serivce Page # ValleyCare Olive View-UCLA Medical Center 14445 Olive View Dr. Sylmar, CA 91342 Pediatric Allergy 92 Pediatric Asthma 93 Pediatric Cardiology 94 Pediatric Cleft Palate 95 Pediatric Clinic Health Centers 96 Pediatric .
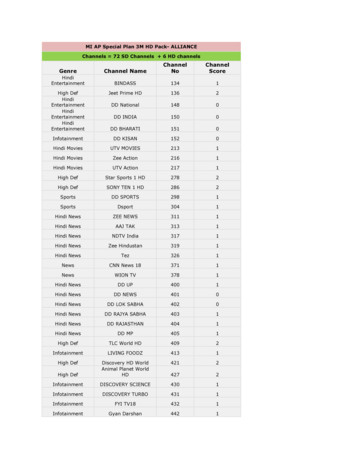
Kids CARTOON NETWORK 449 1 Kids Pogo 451 1 Kids HUNGAMA 453 1 Kids NICK 455 1 Kids Marvel HQ 457 1 Kids DISNEY 458 1 Kids SONIC 460 1 . Hindi News AAJ TAK 313 1 Hindi News NDTV India 317 1 Hindi News News18 India 318 1 Hindi News Zee Hindustan 319 1 Hindi News Tez 326 1 Hindi News CNBC AWAAZ 329 1 .
Coaxial, Multi-Conductor, Paired & Speaker Cable 3 LEVELS OF SAVINGS NAME HOUSE BRANDS BRANDS SPECIAL BUY Wire & Cable 42 Order by Phone at 1-800-831-4242 or Online at www.Jameco.com Coaxial Cable † Color: black † PVC jacket † Polyethylene insulated