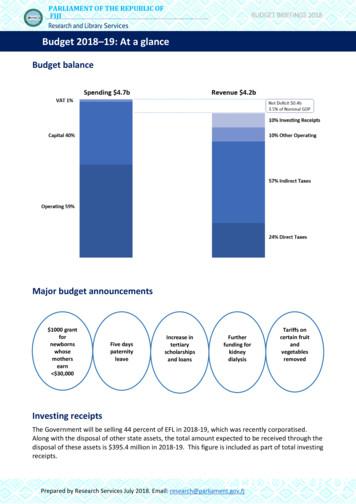
SingleCellNet: A Computational Tool To Classify Single Cell RNA-Seq .
Cell Systems, Volume 9 Supplemental Information SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data Across Platforms and Across Species Yuqi Tan and Patrick Cahan
Supplementary Information Figure S1. Related to Figure 1 SCN parameter and performance correlation. Figure S2. Related to Figure 1 Benchmark performance between SCN and SCMAP Figure S3. Related to Figure 1 Compute time comparisons between SCN and SCMAP Figure S4. Related to Figure 2 Additional analysis on the application of SCN across scRNASeq platforms to determine the identity of unknown cells. Figure S5. Related to Figure 2 Application of SCN in cross-platform and cross-species analysis on PBMCs Figure S6. Related to Figure 2 Application of SCN to compare cortex atlas across studies, Tasic et al adult mouse cortex scRNA-Seq atlas is queried against Zeisel et al, an adult mouse brain atlas, based SCN-TP classifier. Figure S7. Related to Figure 2 Application of SCN to compare cortex atlas across studies, Tasic et al adult mouse cortex scRNA-Seq atlas is queried against Zeisel et al, an adult mouse brain atlas, based SCN-TP classifier. Figure S8. Related to Star Methods Step-by-step R function commands used to build a SCNTP classifier and query data. Table S1. Related to Figure 1 scRNA-Seq training and query data sets used for cross-platform comparison Table S2. Related to Figure 1 scRNA-Seq training and query data sets used for cross-species comparison Table S3. Related to Figure 1 Benchmarking values of sixteen dataset pairs between six methods: SCN-TP, SCN-Base, SCMAP-cosine, SCMAP-pearson, SCMAP-spearman, and SCMAP-cell with k. Table S4. Related to Star Methods This is a list of 12 reference data sets with expression matrix and metadata/sample table, which is downloaded through SingleCellNet GitHub page
Supplementary Figure 1 A Effects of dropout correction on classifier performance 1.00 Effects of Top scoring Pairs used on classifier performance Effects of Top scoring Pairs used on classifier performance 0.75 mean AUPR classifier.method 0.50 rf tsp rf 0.50 classifier.method tsp rf 0.50 0.50 0.50 0.25 0.25 0.25 classifier.method rf classifier.method tsp rf tsp rf 0.50 0.25 0.25 0.25 0.00 0.00 0 0.00 1 Top pair per cell type: topX Feature genes in total: xpairs 0.75 0.75 0.75 Mean AUPR mean AUPR 0.75 0.75 mean AUPR cohen's kappa Cohen’s kappa 1.0 Effects of dropout correction on classifier performance 1.0 1.00 no 1 32 magic dropout.correction 5 10 topX 5 160 10 320 0 yes train only 15 15 480 0.00 Top pair per cell type: topX 11 Feature genes in total: xpairs 32 no magic dropout.correction 5 10 5 160 10 topX 320 yes train only 15 15 480 B 1.0 mean AUPR Cohen’s kappa cohen's kappa 1.00 Effectsof ofcell cellcycle cycle regression classifier performance Effects regression onon classifier performance 0.75 0.75 1.0 Effects of cell cycle regression on classifier performance 1.00 0.75 0.75 mean AUPR Mean AUPR 0.75 0.50 0.50 classifier.method rf tsp rf classifier.method 0.50 rf classifier.method tsp rf 0.50 0.50 0.25 rf tsp rf 0.25 0.25 0.25 0.25 0 0 0.00 0.00 0.00 no no yes yes train only yes query only cell.cycle.regression no yes yes train only yes query only cell.cycle.regression yes yes train only yes query only cell.cycle.regression Figure S1. Related to Figure 1 SCN parameter and performance correlation (A) The Tabula Muris 10x data set cross-validation was used to test how the number of top–pairs used in SCN can influence its performance. We have tested 1 top pair per cell type (32 genes in total), 5 top pairs per cell type (160 genes in total), 10 top pairs per cell type (320 genes in total), and 15 top pair per cell type (480 genes in total). The performance of SCN plateaus at 10 top-pairs per cell type for both mean AUPR and k. (B) How cell cycle regression may affect SCN performance. We have examined four conditions where 1) both training and query data have not been regressed for cell cycle, 2) both training and query data have regressed out cell cycle effect regressed out, 3) only training data has cell cycle effect regressed out. 4) only query data has cell cycle effect regressed out.
(H n ro a b (M n ro a b ) T )( (H n ro a b b ar )( ro a o T )(Q ro n() n( M ) n H ba )(T (H mu )(Q ) ro ( ) T ran ) n( m ) o H urse (Q )(T agn ) ) eo rs (Q se ge ba tolp ) r rs o e( Q ba b ton( ro aro lHp)( e ) n( n T) (Q ba ( H M x ) ro )(T ) i n( )( T) n(Q M )(T ba xbain ) ) ro ro (Q ba ba n( n( ) ro b roM) H) a n( n( ( M ron T() H Q) ba )(T (M m )(Q ro u ) m )(T ra ) n( M ur) no )(T ase n (Q ) g o ) e (Q se ge b rsto ) rsar lp toon e( ba (lMpe Q) ro n( )( (Q T M )(Tt ) x ) m) in 1 tm 0 x (Q 10 t x(T in(Q) x( m1 ) ) tm T) 0 m xm 10 (T ws x( ) w (Q T) m s( ) m w Q w tm s(ra ) s(1 w tm r0 a ) x w (Q 10 x( tm (T) )(Q) tm T) 10 t ) 10 x( mf tm a T x( ) fa cs T) tm cs (Q t fa (Q) m c ) fa cs s(r t(mr a w a tm fa w )( c fa Q cs tm s(T )(Q) (T fa ) ) tm ) c m fa sm cs (T w ws (T ) s (Q ) m (Q) m w t ws( ) sm( ra tm fara w fa w cs t cs( )()(Q Q (T m T) ) ) tm ) fac t fa tsm m cs (T 1 10 (T ) 0x x ) tm ((Q Q tm 10 10 ) ) x( x(r ra aw w) )((Q Q )) ba ) b )(T H n( ro ba Cohen's Kappa Cohen’s Kappa Mean AUPR 0.25 0.25 SCN: Base method SCN: Top pair SCMAP cell SCMAP cluster cosine SCMAP-cluster: Cosine SCMAP cluster pearson SCMAP cluster spearman SCMAP-cluster: Pearson SCN Base SCN TP c1 tm SCMAP-cluster: Spearman ) x( ra w 0 1 ) x 0 1 tm tm c1 ra w s( ) ra w x( ) 0 x 1 tm ra w s w m ) (r a w s( 1 0 tm c1 tm c1 tm w m c1 tm c1 ) c1 tm c1 tm tm x 0 1 tm s( ra w x 1 0 tm s w m ) SCMAP-cluster: Pearson tm w ) s w m c1 m c1 tm tm (r a w c1 tm c1 tm x 0 1 tm x 0 1 ) ra w s( w m 0 x (r a w x 1 0 tm ) c1 tm c1 tm s 1 tm m w s w m SCMAP-cluster: Cosine tm w SCMAP cluster pearson SCMAP cluster spearman m x 0 ) s w m x 0 1 tm (r a w c1 tm c1 tm s 1 tm w m s SCMAP cell SCMAP cluster cosine w x x( ra w 0 1 1 0 tm s tm s w m xi n ) lp to e n o SCN: Top pair m ) ra w x( 0 1 x 0 1 tm s tm s w m w m w (H n a ro b rs g e se m u ra ) SCN: Base m xi n method ) 0.75 (H n ro a (H ) (H n ro a b ro n a b mean AUPRC 0.75 0.25 Mean AUPR A b e o n lp rs to e g se ) (H n 0.75 ra m u 0.75 0.25 Cohen’s Kappa 1.0 ro ) (H 1.00 a b n ro a b cohen.s.kappa Supplementary Figure 2 SCMAP-cluster: Spearman 1.00 1.0 Same species – cross platform 0.75 0.50 0.50 0.25 0.00 0 SCN Base SCN TP dataset 0.50 0.50 1.0 0.25 0.00 0 dataset B0.50 SCMAP-cell 1.0 1.0 0.75 0.75 0.25 0.50 0.50 00 0 dataset dataset Figure S2. Related to Figure 1 Benchmark performance between SCN and SCMAP (A) Fifteen pairs of cross-platform scRNA-Seq training-query data sets were used to benchmark the performance of the five methods: SCN-TP, SCN-Base, SCMAP-cosine, SCMAP-pearson and SCMAP-spearman, using two quantitative metrics: mean AUPR and k. The training for each pair of the cross-platform comparisons was bootstrapped ten times. The barplot exhibits the
classifier performance of all ten training by displaying the average AUPR and standard deviation as error bar. (B) Benchmark sixteen dataset pairs between six methods: SCN-TP, SCN-Base, SCMAP-cosine, SCMAP-pearson, SCMAP-spearman, and SCMAP-cell with k.
method ba scmap cell annotation scmap cell projection scmap cluster s(T dataset SCN ) ) (Q x(Q ) ) (Q (Q (ra w) 0x m1 m1 0 ) t s(T ) t fac tm tm fac (ra w) ws ) ) (Q ) m ws s(T ) m s(T fac tm fac s(Q ) (Q s(r aw ) mf ac ) t x(T mf ac ) t (ra w) SCN cgenes tm tm 10 x(T 10 ws ) m ) (Q ws ) m SCMAP tm x(T 10 Q) in( ) (Q pe ) ) (Q )(Q no ) x )(T x(T tm 10 (M ron tol ers eg ) s ba )(T n(H ) m ura )(T (M method tm (M ron ron ba Q) in( ) (Q pe ) ) (Q no ) x aro ) b (M )(T ron )(T tol ers (H ron eg ) s ba )(T ) m ura )(T )(Q n(M aro log10(system.time) B ba (H ron ba (H ron ba ) b )(T (H ron ba ) t (Q (ra w) (Q ) ws m in( Q) ) ) mf ac 0x s(Q (T) tm ) fac s(r aw )(Q tm fac ) s(T ) m tm fac ws s(T (Q ) ) m ws (ra w) tm (Q fac ) s(T ) t tm m1 fac 0x s(T (Q ) t ) m1 0x (ra w) (Q ) tm 1 x(T 10 tm (T) 0x ) m ws x(T 10 tm tm 1 (Q pe ) (Q no ) x )(T (M ron ba tol ers eg ) s )(T ) Q) )(Q n(H ) m ura )(T aro x in( ) (Q ) (Q pe tol no ) )(Q n(M (T) (H ) ) b )(T ron (M ron (M ron ba ba (M ron ba ba ers eg ) s )(T aro ) m ura (H )(T ron (H ron ba ba ) b )(T (H ron ba log10(system.time) Supplementary Figure 3 A Feature selection time between SCN and SCMAP 3 2 1 0 1 dataset SCN xpairs Classification/projection time between SCN and SCMAP 2 0 2
Figure S3. Related to Figure 1 Compute time comparisons between SCN and SCMAP (A) Feature selection time on 16 dataset-pairs between SCN and SCMAP. SCN has two feature selection steps: calculating the most differentially expressed genes (cgenes) and top-pair selection (xpair). X-axis lists the data sets, Y-axis display system time in log10(s) scale. (B) Classification or projection time on sixteen dataset-pairs between SCN and SCMAP. SCMAPcell has two projection steps: cell-cell projection (projection) and cell annotation (annotation). Xaxis lists the data sets, Y-axis display system time in log10(s) scale. *Only a subset of MWS data set (6477 cells) is used for query in this study (Supp Table 1).
Supplementary Figure 4 SingleCellNet classifier { } Training data: Baron et al. cell type annotation —— —— (—— – –) – – (——) { } Query data: Segerstolpe et al cell type annotation —— —— (—— – –) – – (——) acinar alpha beta co-expr delta ductal Unknown epsilon MHC II EC mast gamma PSC acinar activated stellate alpha beta delta ductal endothelial epsilon gamma macrophage mast quiescent stellate unknown schwann T cell acinar activated stellate alpha beta delta ductal endothelial epsilon gamma macrophage mast quiescent stellate unknown schwann T cell acinar activated stellate 1 0.8 0.6 0.4 alpha 0.2 beta 0 delta ductal endothelial epsilon gamma macrophage mast quiescent stellate rand schwann T cell Figure S4. Related to Figure 2 Additional analysis on the application of SCN across scRNASeq platforms to determine the identity of unknown cells. Baron adult human pancreatic scRNASeq data and its annotation provided by the authors are used to train SCN-TP classifier. Segerstolpe et al profiled adult human pancreatic scRNA-Seq data with a different scRNA-Seq technique was used as query data. A classification heatmap is used to visualize the group unclassified unclassified endoc
classification result of the Segerstolpe data on the Baron-based SCN-TP classifier. The annotated columns are labeled according the provided label from the Segerstolpe group. The row names indicate the cell-type specific classifier trained with the Baron data. The classification score for each column/cell sums up to 1, with a range from 0 (black) to 1 (yellow).
m he s B m tem cel on c l o e N cy ll K te at c op T ell oi ce et ll ic st B e c m m el on c l he oc ell N y m K te at c op T ell oi ce et ll ic B s m tem cel on c l he oc ell N y m K te at c op T ell oi ce et ll ic st B m em cel on c l he o e N cy ll m K te at c op T ell oi ce et ll ic st B m em cel on c l he o e N cy ll m K te at c op T ell oi ce et ll ic st B m em cel on c l e he o N cy ll m K te at c op T ell oi ce et ll ic st B m em cel on c l he oc ell N y m K te at c op T ell oi ce et ll ic s B m tem cel on c l he oc ell N y m K te at c op T ell oi ce et ll ic B s m tem cel on c l he o e N cy ll m K te at c op T ell oi ce et ll ic st B m em cel on c l he o e N cy ll m K te at c op T ell oi ce et ll ic st B m em cel on c l o e N cy ll K te c T ell ce ll ic ie t op o at m he tic ie o p to a Class score Class score Classification score Class score 1.0 group C 0 s s BB e cc m temtm ono m cecll ell no e e h o cl ll e m m NNKcy lyt K te e at a to c ce op p oi oi TT cecll ll et et e el ic ic ll l s s BB e cc m temtm ononm cecll ell he oo cel ell h e m m NNKcy lyt K te e at a to c ce op oi poi TT cecll ll et et el el ic ic l l st st BB e cc m em m ononm cecll ell he h oo el el e m m NNKccy lyt l K te e at a to cece op oi poi TT c cll ll et et el el ic ic l l st st BB e cc m em m ononm cecll ell he h oo el el e m m NNKccy ly l K te te at a t cc op o p TT el ell oi oi c cl et et el el ic ic l l st st BB e e c m m mm ecl el onon c cl l h he oo el el e m NN ccy ly l m KK te te a at to cece op oi poi TT c cll ll et et el el ic ic l l st st BB e cc m em m me onon c cll ell h he oo el el e m NN ccy ly l m KK te te a at to cec op e oi poie TT c cll ll et t el el ic ic l l st steBB m em mm cecl el onon c cl l h he oo el e e m NN ccy ly ll m KK te te a at t cc o op p TT el el oi oie cecl l et ti ll el l ic c s steBB c m ec m temom onn c cll ell h he oNoccell ell e m N yy m KK te te a at to cc op Te e p oi oie Tcecll ll et ti ll el l ic c st steBB c m emomm ecl el onn cecl l h he ooccll ell e N m N yy m KK te te a at to c op T ece p oi oie Tcecll ll et ti ll el ic c B l st steBc m emomm ecll el onn cec l h he oo l e e m NN ccy ly ll m KK c te te a at to op p TT ecl e o oi c l ll et ieti ecll el ic c B l st steBc e m momm ecll el onn cec l oo l e NNKccyt ly ll Kc e te c TT cell ell ece ll l l he m ic e h et oi op at m he —— —— —— —— Supplementary Figure 5 A Training data: TM10x Query data: 10x beads purified B cell tcell cd4 helper tcell cd8 cytotoxic B cell cd34 D 1 1.0 B cell B cell endothelial cell endo 0% granulocyte hematopoietic precursor late cell pro B cell granulocyte HPC SingleCellNet classifier (—— – –) – – – – (——) (——) { } { } 25% late pro-B cell type annotation (—— – –) cell type annotation B Original annotation endothelial cell granulocyte hematopoietic precursor latecell pro B cell macrophage endothelial cell granulocyte hematopoietic precursor latecell pro B cell macrophage macrophage count macrophage cluster monocyte monocyte 50% monocyte monocyte natural killer cell natural killer cell natural killer cell NK cell rand rand 75% rand rand T cell T cell T cell NK cell monocyte hematopoietic stem cell 0.50 T cell T cell trachea epithelial 1.0 rand SCN classification category trachea epithelial B cell endothelial cell granulocyte hematopoietic precursor cell late pro B cell macrophage natural killer cell rand T cell trachea epithelial 0.50 0.5 0.50 0 0 cluster SCN classification cluster 0 cluster trachea mesenchymal B cell B cell hematopoietic stem cell monocyte NK cell T cell 100% trachea epithelial tracheae epi Original annotation cluster cluster B cell hematopoietic stem cell monocyte NK cell T cell B cell hematopoietic stem cell monocyte NK cell T cell
Figure S5. Related to Figure 2 Application of SCN in cross-platform and cross-species analysis (A) The hematopoietic-lineage related subset of the Tabula Muris 10x scRNA-Seq data and its annotations are used as training data. The bead-purified 10x sequencing of the human hematopoietic lineage cells are used as query. (B) The classification heatmap shows that human B cells, monocytes, nature killer cells, CD4 T cells and CD8 T cells are well-classified by the mouse TP-RF classifier. (C) This cross-platform and cross-species analysis can also be summarized with an attribution plot, showing that despite some cross-classification signals due to the similarity, most human hematopoietic cells are accurately classified. (D) The classification score is visualized with violin plot, where x-axis shows the annotation of the query cells provided in the original studies, and y-axis is the range of classification score in each given category. endo: endothelial cell; HPC: hematopoietic precursor cell; late pro-B: late pro-B cell; NK cell: natural killer cell; tracheae epi: tracheae epithelial cell.
Supplementary Figure 6 SingleCellNet classifier { } { } Training data: Zeisel cell type annotation —— —— (—— – –) – – (——) Query data: Tasic cell type annotation —— —— (—— – –) – – (——) Astroependymal cells 1.0 0.50 0 Cholinergic, monoaminergic and peptidergic neurons 1.0 0.50 0 Di and mesencephalon neurons 1.0 0.50 0 Hindbrain neurons 1.0 0.50 0 Immature neural 1.0 Original annotation cluster 0.50 Astrocyte Endothelial cell GABA ergic neuron Glutamatergic neuron Microglia Oligodendrocyte Oligodendrocyte precursor cell Unclassified Immune cells 1.0 0.50 0 Oligodendrocytes 1.0 0.50 0 Spinal cord neurons 1.0 0.50 0 Telencephalon interneurons 1.0 0.50 0 Telencephalon projecting neurons 1.0 0.50 0 Vascular cells 1.0 0.50 dr en od lig d si fie nc U ec ur pr e oc yt O O la s so oc dr en od lig er SCNcluster classification cluster rc el l yt e lia M ne c gi ic ro g on ur ur at m ta G lu G AB A En er do th gi c el ne ia l cy ce te ll on 0 As tro Classification score Class score 0
Figure S6. Related to Figure 2 Application of SCN to compare cortex atlas across studies. Tasic et al adult mouse cortex scRNA-Seq atlas is queried against Zeisel et al, an adult mouse brain atlas, based SCN-TP classifier. The classification score is visualized with a violin plot, where x-axis shows the annotation of the query cells provided in the original studies, and y-axis is the range of classification score in that combination, and the plot is then faceted by the classifier categories.
Supplementary Figure 7 A Original annotation B Unclassified C 100% 100% Unclassified Oligodendrocyte precursor cell Oligodendrocyte precursor cell 75% Oligodendrocyte 100% 100% 75% Unclassified Oligodendrocyte 75% 75% group Oligodendrocyte count group Microglia Glutamatergic neuron 50% 50% count Glutamatergic neuron 50% 50% group Microglia count Oligodendrocyte precursor cell Microglia Glutamatergic neuron GABA ergic neuron 25% GABA ergic neuron GABA ergic neuron 25% 25% 25% Endothelial cell Endothelial cell Endothelial cell Astrocyte 0% 0% 50% 75% 0% 100% d 25% unclassified nc la 0% ss ifie Astrocyte U Astrocyte Cholinergic, monoaminergic and peptidergic neurons 25% 50% 75% Di and mesencephalon neurons si Telencephalon projecting neurons Immature neural Spinal cord neurons Vascular cells Immune cells count Telencephalon interneurons as Oligodendrocytes 100% fie d group Hindbrain neurons group Astroependymal cells category SCN classification Astroependymal cells count Cholinergic, monoaminergic and peptidergic neurons Di and mesencephalon neurons Hindbrain neurons Hindbrain neurons Oligodendrocytes Immature neural Spinal cord neurons U nc la 0% 0% 100% U Astroependymal cells 75% nc l 50% count ed 25% ss ifi 0% Immune cells Oligodendrocytes Telencephalon interneurons Telencephalon projecting neurons Vascular cells Cholinergic, monoaminergic and peptidergic neurons Immature neural Spinal cord neurons Di and mesencephalon neurons Immune cells Telencephalon interneurons Telencephalon projecting neurons group Vascular cells
Figure S7. Related to Figure 2 Application of SCN to compare cortex atlas across studies. Tasic et al adult mouse cortex scRNA-Seq atlas is queried against Zeisel et al, an adult mouse brain atlas, based SCN-TP classifier. (A) The classification heatmap displays the classifier categories in rows, and the original cell label is clustered and colored at the top of column. (B) A general attribution plot summarizes the composition of classification within each label groups. (C) A zoom-in attribution plot of the previous unknown category in the Tasic et al data shows that the majority of the unknown cell types are mostly microglia, astroependymal cells, cholinergic, monoaminergic and peptidergic neurons, and immune cells.
Supplementary Figure 8 I. Building the classifier II. Classifying query data R Functions R Functions Step 0 If (Species conversion TRUE) a. csRenameOrth() Step 1 Subset training data a. splitCommon() Step 2 Pair transform a. findClassyGenes() b. ptGetTop() Gene A Genes 0132010 1020201 0103130 3011322 Gene B a1 b1 c1 c2 b2 a2 b3 Gene pairs Genes a1 b1 c1 c2 b2 a2 b3 a1 b1 c1 c2 b2 a2 b3 0111010 1000010 0100101 1010101 0011000 0101010 Step 6 Template matching a. query transform() Step 3 Template matching a. query transform() precision a1 b1 c1 c2 b2 a2 b3 1000010 0100101 0011000 Step 4 Train classifier a. sc makeClassifier() C B recall Cohen’s kappa mean AUPRC a. b. c. d. e. Step 5 assess query transform() rf classPredict() assess comm() plot PRs() plot metrics() Gene pairs Gene pairs a1 b1 c1 c2 b2 a2 b3 A 0132010 1020201 0103130 3011322 1000010 0100101 0011000 Step 7 Classifying query a. rf classPredict() a. b. c. d. e. Step 8 visualization sc hmClass() plot umap() plot attr() skylineClass() sc violinClass()
Figure S8. Related to Star Methods Step-by-step R function commands used to build a SCNTP classifier and query data. The detailed functionality for each step is elaborated in the Method Section.
Table S1. Related to Figure 1: scRNA-Seq training and query data sets used for crossplatform comparison Cross-platform Training Data Query Data Baron Murano, 2120 adult human pancreatic cells, 8596 adult human CEL-Seq2 same species analysis 1 pancreatic cells, inDrop 2 Segerstolpe, 2209 adult human pancreatic cells, Smart-Seq2 3 4-5 6–7 Xin, 1492 adult human pancreatic cells, C1 MWS, TM10x TM10x (raw) tabula muris 10x data, 6477 adult mouse cells 24936 adult mouse cells across 32 cell types, across 125 cell types, 10x Microwell-seq TMfacs TMfacs (raw) tabula muris facs data, 40620 adult mouse cells across 69 cell types, smartseq2 8-9 10 - 11 TM10x, MWS MWS (raw), 6477 mouse cells across 1599 adult mouse cells 125 cell types, Microwell-seq across 32 cell types, 10x TMfacs TMfacs (raw) tabula muris facs data, 40620 adult mouse cells across 69 cell types, smartseq2 12 - 13 14 - 15 TMfacs, MWS MWS (raw), 6477 adult mouse cells 3182 adult mouse cells across 125 cell types, Microwell-Seq across 69 cell types, smartseq2 TM10x TM10x (raw) (tabula muris 10x data), 24936 adult mouse cells across 32 cell types, 10x *(raw) indicates the query expression matrix was used in raw count form
Table S2. Related to Figure 1: scRNA-Seq training and query data sets used for cross-species comparison Cross-platform cross-species analysis Training Data Query Data 1 Baron 1886 adult mouse pancreatic cells, inDrop Murano, 2120 adult human pancreatic cells, CEL-Seq2 2 Segerstolpe, 2209 adult human pancreatic cells, Smart-Seq2 3 Xin, 1492 human pancreatic cells, C1 4-5 MWS 4038 adult mouse brain cells, Microwell-Seq Darmanis Darmanis (raw), 331 adult human brain cells, C1 *(raw) indicates the query expression matrix was used in raw count form
Table S3. Related to Figure 1 Benchmarking values of sixteen dataset pairs between six methods: SCN-TP, SCN-Base, SCMAP-cosine, SCMAP-pearson, SCMAP-spearman, and SCMAP-cell with k. () in SCMAP-cell column denotes the performance assessment of SCMAPcell when only cells with assignment were evaluated. Kappa SCMAP- SCMAP- SCMAPDataset SCN-TP SCN- cluster- cluster- cluster- base cosine pearson spearman SCMAP-cell tm10x(T) MWS(raw)(Q) 0.631 0.408 0.647 0.653 0.634 0.031 (0.19) tm10x(T) tmfacs(Q) 0.785 0.817 0.749 0.773 0.787 0.153 (0.294) tm10x(T) tmfacs(raw)(Q) 0.772 0.811 0.188 0.186 0.838 0 (0.172) tm10x(T) MWS(Q) 0.631 0.669 0.626 0.663 0.643 0.08 (0.259) tmfacs(T) tm10x(Q) 0.713 0.742 0.667 0.69 0.687 0.24 (0.386) tmfacs(T) tm10x(raw)(Q) 0.706 0.748 0.533 0.473 0.659 0.018 (0.6) tmfacs(T) MWS(Q) 0.32 0.395 0.406 0.365 0.343 0.021 (0.251) tmfacs(T) MWS(raw)(Q) 0.332 0.232 0.423 0.307 0.337 0.004 (0.219) baron(M)(T) baron(H)(Q) 0.599 0.824 0.68 0.659 0.439 0.357 (0.687) baron(M)(T) Xin(Q) 0.619 0.575 0.591 0.56 0.206 0.04 (0.657) baron(M)(T) Segerstolpe(Q) 0.794 0.898 0.916 0.895 0.639 0.411 (0.7) baron(M)(T) Murano(Q) 0.827 0.652 0.778 0.729 0.507 0.137 (0.467) baron(H)(T) baron(M)(Q) 0.559 0.881 0.88 0.886 0.597 0.344 (0.927) baron(H)(T) Segerstolpe(Q) 0.903 0.289 0.795 0.879 0.948 0.315 (0.911) baron(H)(T) Murano(Q) 0.766 0.894 0.926 0.903 0.451 (0.842) 0.911 0.205 baron(H)(T) Xin(Q) 0.966 0.673 0.982 0.987 0.897 (0.928)
Table S4. Related to Star Methods This is a list of 12 reference data sets with expression matrix and metadata/sample table, which is downloaded through SingleCellNet GitHub page. * denotes these are just a subset of the original data that can be used directly for training due to the memory limitation of R. Reference datasets Species Murano Cell/ Tissue/Organ type Cell number and Sequencing technology Adult human pancreatic cells, CEL-Seq2 2120 cells, CEL-Seq2 Adult human brain cells 331 cells, C1 Baron Adult human pancreatic cells 8596 cells, inDrop MWS Mouse cells across 125 cell types, 181755 cells, Microwellseq TMfacs tabula muris facs data Adult mouse cells across 69 cell types 40620 cells, Smartseq2 TM10x tabula muris 10x data Adult mouse cells across 32 cell types 24936 cells, 10x Adult mouse pancreatic cells 1886 cells, inDrop Gokce adult mouse striatum cells 1208 cells, C1 and Smartseq2 Haber Adult mouse intestine epithelium 7216 cells, Smartseq2 Park Adult mouse kidney cells 43745 cells, 10x Zeisel Adult mouse brain atlas 1944 cells*, 10x Loo Developing cortex atlas 10931 cells, Dropseq Darmanis Baron Human Mouse
Muris 10x data set cross-validation was used to test how the number of top-pairs used in SCN can influence its performance. We have tested 1 top pair per cell type (32 genes in total), 5 top pairs per cell type (160 genes in total), 10 top pairs per cell type (320 genes in total), and 15 top pair per cell type (480 genes in total).
e Adobe Illustrator CHEAT SHEET. Direct Selection Tool (A) Lasso Tool (Q) Type Tool (T) Rectangle Tool (M) Pencil Tool (N) Eraser Tool (Shi E) Scale Tool (S) Free Transform Tool (E) Perspective Grid Tool (Shi P) Gradient Tool (G) Blend Tool (W) Column Graph Tool (J) Slice Tool (Shi K) Zoom Tool (Z) Stroke Color
6 Track 'n Trade High Finance Chapter 4: Charting Tools 65 Introduction 67 Crosshair Tool 67 Line Tool 69 Multi-Line Tool 7 Arc Tool 7 Day Offset Tool 77 Tool 80 Head & Shoulders Tool 8 Dart/Blip Tool 86 Wedge and Triangle Tool 90 Trend Fan Tool 9 Trend Channel Tool 96 Horizontal Channel Tool 98 N% Tool 00
theoretical framework for computational dynamics. It allows applications to meet the broad range of computational modeling needs coherently and with fast, structure-based computational algorithms. The paper describes the SOA computational ar-chitecture, the DARTS computational dynamics software, and appl
work/products (Beading, Candles, Carving, Food Products, Soap, Weaving, etc.) ⃝I understand that if my work contains Indigenous visual representation that it is a reflection of the Indigenous culture of my native region. ⃝To the best of my knowledge, my work/products fall within Craft Council standards and expectations with respect to
computational science basics 5 TABLE 1.2 Topics for Two Quarters (20 Weeks) of a computational Physics Course.* Computational Physics I Computational Physics II Week Topics Chapter Week Topics Chapter 1 Nonlinear ODEs 9I, II 1 Ising model, Metropolis 15I algorithm 2 Chaotic
Adobe InDesign Photoshop Move tool Marquee tool - for selection Lasso tool - free form for selection Magic Wand tool - for color selection Crop tool Eyedropper tool - to get color from document Spot Healing tool - to get rid of blemishes Brush tool - paintbrush and pencil Stamp tool - for using textures in your document to paint over other areas
you tried?" gives children the competence and confidence to solve their own problems. Every Tool can be communicated through embodied hand-gestures and modeling by adults. & The 12 Tools . Breathing Tool Quiet/Safe Place Tool Listening Tool Empathy Tool Personal Space Tool Using Our Words Tool Garbage Can Tool Taking Time Tool Please & Thank .
MI6 adventure, Alex Rider is recruited right off the soccer field to check out some suspicious goings-on at Wimbledon. This assignment catapults him into a series of life-threatening episodes, such as coming face to face with a great white shark, dodging bullets as he dives off a burning boat, and being tied to a conveyor belt that is moving toward the jaws of a gigantic grindstone in an .