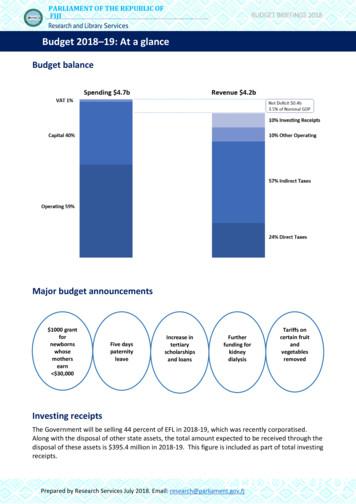
MtGenome Sequencing Of NIST Reference Materials And .
mtGenome Sequencing of NISTReference Materials andPopulation SamplesKevin Kiesler, M.S.Research Biologist - NIST Applied Genetics GroupISHI Workshop: The Future is Now for MPS mtDNA AnalysisSeptember 24, 2018
Outline Reference materials and mtDNA sequencing Population scale sequencing Informatics
First – the Disclaimer Disclaimer - Points of view in this document are those of the authors anddo not necessarily represent the official position or policies of the U.S.Department of Commerce or the Department of Justice. Certaincommercial equipment, instruments, and materials are identified in orderto specify experimental procedures as completely as possible. In no casedoes such identification imply a recommendation or endorsement by NIST,nor does it imply that any of the materials, instruments, or equipmentidentified are necessarily the best available for the purpose. All work presented has been reviewed and approved by the NIST HumanSubjects Protections Office.
What is this?A) Time machineB) Water purifierC) Steam locomotiveD) Mass spectrometer
What is this?Sample plates stack hereThe PLEX-IDFrom Abbott Molecular& Ibis BiosciencesThe mass spectrometersits inside hereSample purificationgoes on inside hereNIST evaluated thissystem in 2012 forpotential “rapid, simple”mtDNA analysis
mtDNA by Mass Spectrometry PCR across HV1/HV2/HV3 Eight tri-plex reactions 24 amplicons
mtDNA by Mass Spec Desalt PCR products (automated) Electrospray injected Time-of-Flight (ESI-TOF) mass spectrometer Review data for “trouble” Sodium adducts, overlapping peaks, etc. Compare to database Known mtDNA types Masses of PCR products Predict DNA sequence Paper: compared with Sanger n 711 samples 99.97 % concordantOutcome:Simplified, faster workflowAlmost as informative as Sanger
NIST Standard Reference Materials (SRMs) ISO/Guide 30:2015 Reference materials (RMs) and certified reference materials (CRMs) (defined in 2.1 and 2.2)are widely used for the calibration of measuring apparatus, for the evaluation ofmeasurement procedures and for the internal or external quality control of measurementsand laboratories. Assess a new technology - calibration Evaluate a method - implementation
NIST Mitochondrial Sequencing SRMs SRM 2392 Three components Component A: DNA from cell line CHR Component B: DNA from cell line 9947A Component C: Cloned fragment from HV1 region of CHR containing C-stretch SRM 2392-I One component DNA from cell line HL60 Characterized with Sanger methods Released in 2001
Assessing NGS Using Reference Materials In 2012, all we had was a SOLiD system Sequencing by Oligonucleotide Ligation and Detection (SOLiD) Ligation chemistry based (it’s complicated) system for whole human genomes Very high coverage mtGenome sequence 45,000 ation-sequencing-technologies
All the Sanger-Based Information Checked OutThere was no software available thatcorrectly handled these C-stretchinsertions with forensic nomenclature. XXX Reference material Technology
We Could Detect Some New Features! Heteroplasmy below what we can see with Sanger methodsSRM 2392 Component A (CHR)Position 1,393 R (17 % A)SRM 2392-I (HL-60)Position 2,445 Y (7 % C)
We Sent Some Samples to Service Labs, Friends Beckman Genomics sequencing service Seven mtGenomes One lane on a HiSeq 40,000 X coverage!!! ( 3 to 4 million bases each) Too much to display in IGV (not enough RAM) Edge Bioserv (Gaithersburg, MD) Ion Torrent Personal Genome Machine (PGM) in 2013 Seven mtGenomes on a 314 chip 200 X to 400 X coverage Illumina MiSeq in 2013 Seven mtGenomes on 2 x 150 v2 chemistry 10,000 X to 20,000 X coverage Children’s National Hospital (in collaboration) Pacific Biosystems RS in 2014 Long-read technology, low coverage Some variants missed
Then We Got Some Desktop Sequencers ofOur Own Ion Torrent PGM September 2012 Chemistry improvements over time 200 bp, 300 bp, 400 bp, HiQ Illumina MiSeq September 2013 Chemistry improvements over time V2 2x150 bp, V2 2x250 bp, V3 2x300 bp
Multiple Orthogonal Measurements Great approach for certifying reference materials!SRM 2392ComponentANucleotide64 TPGM Edge26.8PGM NIST 124.3PGM NIST 225.0PGM NIST 329.7PGM NIST HiQ33.2MiSeq Edge33.0MiSeq NIST31.6HiSeq BC30.6SOLiD NIST29.0Average29.2St. Dev.3.1Component B1393 A7861 7.988.416.988.316.787.317.474.61.714.5SRM .89.16.47.47.17.37.08.47.31.31.1
Multiple Orthogonal Measurements Great approach for characterizing reference materials!SRM 2392ComponentANucleotide64 TPGM Edge26.8PGM NIST 124.3PGM NIST 225.0PGM NIST 329.7PGM NIST HiQ33.2MiSeq Edge33.0MiSeq NIST31.6HiSeq BC30.6SOLiD NIST29.0Average29.2St. Dev.3.1Component B1393 A7861 7.988.416.988.316.787.317.474.614.51.7It can also educate you about your technology.SRM .89.16.47.47.17.37.08.47.31.31.187 %
Multiple Orthogonal Measurements Great approach for characterizing reference materials! SOFTWARE
Conclusions Reference materials Can identify technical limitations/bias Often need multiple measurements Orthogonal techniques Help to select best procedures
Population Scale Sequencing
Project Planning - Goals Submit forensic-quality whole mtGenome data to EMPOP Current database (V3, Release 11) n 26,127 control region sequences n 256 whole genome sequences NIST population samples (n 1,000) African American, Asian, Caucasian, Hispanic Sequencing plan Start with Caucasian population 440 mtGenomes
Project Planning Questions What instrument do we use? What protocol/chemistry do we use? What analysis procedure do we use? Software, data review, etc.Illumina MiSeq FGxIon Torrent S5Ion Chef
Project Planning: Instrument Selection Considerations Cost Time/laborCost Per 96 Samples (Approximate) 30,000 25,000 Automation 20,000 15,000 10,000 5,000 -S5XL AutomatedLibrary PrepS5XL ManualLibrary PrepMiSeq ILMNMethodMiSeq AFDILMethod
Project Planning: Protocol Selection Options Illumina whole genome procedure Long PCR primers developed by Dr. Mark Wilson’s labTaKaRa LA TaqIllumina Nextera XT library preparationIllumina MiSeq v2 2x150 cartridge (per protocol) AFDIL whole genome procedure Long PCR primers from Fendt et al., BMC Genomics 2009, 10:139TaKaRa LA Taq (GC Buffer & BSA)Kapa HyperPlus Library KitIllumina V3 2x300 cartridge
Pilot Data – Illumina Procedure AmplificationAmplicon 1 – First try PCR could be more robust1 2 3Failed Not a sample quality issueFailedNTC Only one of two amplicons affected4 5 678 9 10 11 SSAmplicon 1 – Second try (re-PCR)1 2 3Poor 10 kbPoor Reproducible 10 kbNTC 18 of 95 ( 18 %) of samples - low yield4 5 6 7 8 9 10 11 SSSS Size standard
Pilot Data – Illumina Procedure AmplificationAmplicon 1 – First try PCR could be more robustNTC Amplicon 1 primer binding site mutation 16 of 18 low/neg samples had this All haplogroup T (G1,888A) HG-T frequency 10 % in Europeans1 2 3Failed Reproducible 10 kbFailed 18 of 95 ( 18 %) of samples - low yield4 5 678 9 10 11 SSAmplicon 1 – Second try (re-PCR)AACTTTGCAAGGAGAGCCAAPrimer binding site1 2 3PoorPoorNTC 10 kb4 5 6 7 8 9 10 11 SSSS Size standard
Library QC – Illumina Procedure Nextera XT library Default parameters per protocol 1 ng DNA 5 minute fragmentation 760 bp
Sequencing Results - Illumina Most samples had 99 % of mgGenome covered Due to sensitivity of Nextera XT library prep & Normalization of PCR input (amplicons 1 & 2 separately) Some gaps in coverage Where amplification was poorGap areaShaded greyGap area
Sequencing Results - Illumina Some gaps in coverage Frequent gaps in HV2 C-stretch Even with good overall coverage
Pilot Data – AFDIL Procedure Amplification Amplicon B gives higher yield than Amp A Amp B contains control regionNTC Primers from Fendt et al. 2009 TaKaRa LA Taq w/GC Buffer & BSA 100 % of samples amplifiedAmplicon A1 2 3 Both amplicons 8 kb4 5 678 9 10 11 SS78 9 10 11 SSAmplicon BNTC Advantage of GC Buffer & BSA?1 2 3SS Size standard4 5 6
Protocol Optimization – Kapa HyperPlus What are the best parameters for library preparation? Fragmentation time 5 min to 30 min Input DNA 10 ng to 1000 ng (PCR product) Optimum library size 2 x 300 bp cartridge planned 450 bp library molecules 300 bp insert 150 bp adaptors No more than 1500 bp poor clusteringKapa HyperPlus Technical Data Sheet (KR1145 v3.16)
Fragmentation TimeOptimizationAll reactions500 ng DNAinput10 minTarget size 450 bp20 min30 min
DNA InputOptimizationAll reactions10 minfragmentation10 ng 450 bp100 ng500 ng
Optimized Conditions Fragmentation time 15 minutes DNA input 300 – 400 ngSome residual adapter dimerTarget size450 bp
Half ReactionTrialAll reactions10 min frag.,500 ng DNAFullRx 450 bpHalfRxReaction kinetics not equal at half volume library construction.
Pilot Data – More Consistent Coverage DepthAFDIL method allows higher multiplexing with less likelihood of dropout sitesIllumina Whole mtGenome Method (Nextera Library Kit)AFDIL Whole mtGenome Method (Kapa Hyper Plus Library Kit)
Pilot Data – Haplogroup Estimation No surprise MT97122MT97123MT97124MT97125Missing 6183C -309.1C onenoneT16093CnonePrivate MutationsT16189CnoneT16189CC10933T A15467GC198T A9327G A13801G T15670CnoneA11252GG709A C9727TG8027A G15301AC16111T T152C-309.1C G4655AT2416C A8817GC3388A C8788TT4373C T15313CG16474TnoneA16138G A73GnoneT16093C G7762AnoneA16158G-309.1C -524.3A I1a1bK1a1b1H1b1H1H2a2aU5a1a1H1 entEurope (H)EuropeEuropeEuropeEurope (H)EuropeEurope (H)Europe (I1a1)EuropeEuropeEuropeEuropeEuropeEuropeEurope (I)EuropeEuropeEuropeEuropeEuropeEurope (T1a)EuropeEurope (U5a)
Half Reaction Library Prep - Save ? Optimized fragmentation conditions DNA input 350 ng Full reaction 15 min Half reactionFull Reaction 20 minTarget size 450 bpHalf Reaction
Compare Results – Full Reaction vs Half No major bias in coverage introduced Half reaction always higher where amplicons 46261495115276156011592616251Difference in Coverage (Full Rx - Half Rx) for Three SRMs Coverage200010000-1000-2000-3000-4000-5000Genome Position2392-A2392-B2392-I
Compare Results – Full Reaction vs Half No major bias in coverage introduced Half reaction always higher where amplicons overlapFull reactionHalf 146261495115276156011592616251Difference in Coverage (Full Rx - Half Rx) for Three SRMs Coverage200010000-1000-2000-3000-4000-5000Genome Position2392-A2392-B2392-I
Compare Results – Full Reaction vs Half Variant sites match 100 %Sample Comparison Tool – GeneMarker HTSFullHalfFullHalfFullHalf
Mixture Sample (3:1) Matches Expected RatioSample Comparison ToolFull reactionHalf reaction
Surprise! Some Degraded Samples Did not amplify with long PCR ( 8 kb) Buccal swabs extracted 2005 Works great with STR multiplexesPositive ControlAmplicon B – No ProductsAmplicon A – No Products1234567891011SSSS Size standard
Thermo Fisher Precision ID Panels Two mtDNA products Whole Genome Panel Two primer pool multiplex 81 primer pairs x 2 pools 162 amplicons 160 bp amplicon size Control Region Panel Two primer pool multiplex7 primer pairs x 2 pools 14 amplicons1.2 kb control region of mtGenome 150 bp amplicon sizeImage credit:
Sequencing Results – Whole Genome Full genome coverage All expected variants detected for SRM 2392 A & B, 2392-I Looking forward to using Thermo Fisher’s analysis software
Sequencing Results – Control Region All expected variants detected for SRM 2392 A & B, 2392-I
Informatics Forensic mtDNA nomenclature is challenging! We should clone Dr. Parson Commercial software Softgenetics GeneMarker HTS Capability for forensic nomenclature EMPOP compatibility CLC Genomics Workbench AFDIL / Qiagen – developed AQME Tool ThermoFisher Scientific Converge mtDNA Analysis (coming soon)
Phylogenetic Analysis – Additional QC StepFor high-throughputgenotyping we willcheck these privatemutation 7121MT97122MT97123MT97124MT97125Missing 6183C -309.1C onenoneT16093CnonePrivate MutationsT16189CnoneT16189CC10933T A15467GC198T A9327G A13801G T15670CnoneA11252GG709A C9727TG8027A G15301AC16111T T152C-309.1C G4655AT2416C A8817GC3388A C8788TT4373C T15313CG16474TnoneA16138G A73GnoneT16093C G7762AnoneA16158G-309.1C -524.3A I1a1bK1a1b1H1b1H1H2a2aU5a1a1H1 entEurope (H)EuropeEuropeEuropeEurope (H)EuropeEurope (H)Europe (I1a1)EuropeEuropeEuropeEuropeEuropeEuropeEurope (I)EuropeEuropeEuropeEuropeEuropeEurope (T1a)EuropeEurope (U5a)
Conclusions Mitochondrial Genome Protocol Multi-factorial decision process for selection Selected AFDIL-developed procedure for reference quality samples Even coverage Higher multiplexing Cost saving with ½ reaction Degraded samples will need a different procedure Analysis method must be high-throughput High accuracy required for EMPOP submission
Save the Date! November 7 Keynote: John ButlerForensic GeneticsFingerprintsDigital & MultimediaFootweat Impression November 8 Keynote: Sheila WillisTrace EvidenceDrugs and ToxinsFirearms and Tool Marks
Thank You!Questions?Contact info:Kevin.Kiesler@NIST.gov Funding Acknowledgements Armed Forces DNA Idenification Laboratory (AFDIL)Kim AndreaggiCharla MarshallThermo Fisher ScientificMatt GabrielNIST Applied Genetics Group NGS TeamDr. Peter Vallone, Group leaderLisa BorsukSarah RimanBecky SteffenKatherine Gettings NIST Special Programs Office: Forensic DNA FBI Biometrics Center of Excellence: Forensic DNA Typing as a Biometric tool.
TaKaRa LA Taq Illumina Nextera XT library preparation Illumina MiSeq v2 2x150 cartridge (per protocol) AFDIL whole genome procedure Long PCR primers from Fendt et al., BMC Genomics 2009, 10:139 TaKaRa LA Taq (GC Buffer & BSA)
2.1 NIST SP 800-18 4 2.2 NIST SP 800-30 4 2.3 NIST SP 800-34 4 2.4 NIST SP 800-37 4 2.5 NIST SP 800-39 5 2.6 NIST SP 800-53 5 2.7 NIST SP 800-53A 5 2.8 NIST SP 800-55 5 2.9 NIST SP 800-60 5 2.10 NIST SP 800-61 6 2.11 NIST SP 800-70 6 2.12 NIST SP 800-137 6 3 CERT-RMM Crosswalk of NIST 800-Series Special Publications 7
NIST SP 800-30 – Risk Assessment NIST SP 800-37 – Risk Management Framework NIST SP 800-39 – Risk Management NIST SP 800-53 – Recommended Security Controls NIST SP 800-53A – Security Control Assessment NIST SP 800-59 – National Security Systems NIST SP 800-60 – Security Category Mapping NIST
NIST Risk Management Framework 1. Categorize information system (NIST SP 800-60) 2. Select security controls (NIST SP 800-53) 3. Implement security controls (NIST SP 800-160) 4. Assess security controls (NIST SP 800-53A) 5. Authorize information system (NIST SP 800-37) 6. Monitor security controls (NIST SP 800-137) Source: NIST CSRC, http .
Source: 9th Annual API Cybersecurity Conference & Expo November 11-12, 2014 - Houston, TX. 11 Industry Standards and Committee Initiatives WIB M2784-X-10 API 1164 ISA 99/IEC 62443 NIST SP 800-82 NIST SP 800-12 NIST SP 800-53 NIST SP 800-53A NIST SP 800-39 NIST SP 800-37 NIST SP 800-30 NIST SP 800-34 ISO 27001,2 ISO 27005 ISO 31000
Also referred to as Next-Generation Sequencing Parallelize the sequencing process, producing thousands or millions of sequences concurrently Lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods. In ultra-high-throughput sequencing as many as 500,000 sequencing-by-synthesis operations may
Mar 01, 2018 · ISO 27799-2008 7.11 ISO/IEC 27002:2005 14.1.2 ISO/IEC 27002:2013 17.1.1 MARS-E v2 PM-8 NIST Cybersecurity Framework ID.BE-2 NIST Cybersecurity Framework ID.BE-4 NIST Cybersecurity Framework ID.RA-3 NIST Cybersecurity Framework ID.RA-4 NIST Cybersecurity Framework ID.RA-5 NIST Cybersecurity Framework ID.RM-3 NIST SP 800-53
Apr 08, 2020 · Email sec-cert@nist.gov Background: NIST Special Publication (SP) 800-53 Feb 2005 NIST SP 800-53, Recommended Security Controls for Federal Information Systems, originally published Nov 2001 NIST SP 800-26, Security Self-Assessment Guide for IT Systems, published Dec 2006 NIST SP 800-53, Rev. 1 published July 2008 NIST SP 800-53A, Guide for
Zoology Practical Manual EM 18-03-2019.indd 7 22-03-2019 11:14:18. 8 5. ABO BLOOD GROUPS - DEMONSTRATION EXPERIMENT AIM: To find out the blood group of a classs / school students. MATERIAL REQUIRED: 1. Human blood sample 5. Spirit (70% alcohol) 2. Antisera A 6. , slides. Lancet 3. Antisera B 7. Cotton 4. Antisera D 8. Mixing sticks PRINCIPLE: The determination of ABO blood group is based on .